-Search query
-Search result
Showing 1 - 50 of 1,792 items for (author: gui & l)

EMDB-18313: 
Retron-Eco1 filament with ADP-ribosylated Effector (local map with 1 segment)
Method: single particle / : Carabias del Rey A, Montoya G

EMDB-18314: 
Retron-Eco1 filament with inactive effector (E106A, 2 segments)
Method: single particle / : Carabias del Rey A, Montoya G

EMDB-18315: 
Retron-Eco1 filament with ADP-ribosylated Effector (full map with 2 segments)
Method: single particle / : Carabias del Rey A, Montoya G

EMDB-18317: 
Retron-Eco1 filament (2 segments)
Method: single particle / : Carabias del Rey A, Montoya G

EMDB-19792: 
Retron-Eco1 -1 turn mutant filament with ADP-ribosylated Effector (Consensus refinement)
Method: single particle / : Carabias del Rey A, Montoya G, Pape T

EMDB-19793: 
Retron-Eco1 filament with ADP-ribosylated Effector (Consensus refinement)
Method: single particle / : Carabias del Rey A, Montoya G

PDB-8qbk: 
Retron-Eco1 filament with ADP-ribosylated Effector (local map with 1 segment)
Method: single particle / : Carabias del Rey A, Montoya G

PDB-8qbl: 
Retron-Eco1 filament with inactive effector (E106A, 2 segments)
Method: single particle / : Carabias del Rey A, Montoya G

PDB-8qbm: 
Retron-Eco1 filament with ADP-ribosylated Effector (full map with 2 segments)
Method: single particle / : Carabias del Rey A, Montoya G

EMDB-41271: 
Consensus map of 96nm repeat of human respiratory doublet microtubule, RS1-2 region
Method: single particle / : Gui M, Brown A

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10
Method: single particle / : Huang J, Ozorowski G, Ward AB

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10
Method: single particle / : Huang J, Ozorowski G, Ward AB

EMDB-36672: 
Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab
Method: single particle / : Liu B, Liu HH, Han P, Qi JX

PDB-8jva: 
Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab
Method: single particle / : Liu B, Liu HH, Han P, Qi JX

EMDB-36366: 
Cryo-EM structure of Symbiodinium photosystem I
Method: single particle / : Zhao LS, Wang N, Li K, Zhang YZ, Liu LN

PDB-8jjr: 
Cryo-EM structure of Symbiodinium photosystem I
Method: single particle / : Zhao LS, Wang N, Li K, Zhang YZ, Liu LN

EMDB-42779: 
Cryo-EM structure of a bacterial nitrilase filament with a covalent adduct derived from benzonitrile hydrolysis
Method: helical / : Aguirre-Sampieri S, Casanal A, Emsley P, Garza-Ramos G

PDB-8uxu: 
Cryo-EM structure of a bacterial nitrilase filament with a covalent adduct derived from benzonitrile hydrolysis
Method: helical / : Aguirre-Sampieri S, Casanal A, Emsley P, Garza-Ramos G
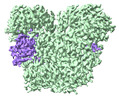
EMDB-36980: 
Cryo-EM structure of DSR2-TTP
Method: single particle / : Zhang H, Li Z, Li XZ

EMDB-36982: 
Cryo-EM structure of DSR2-DSAD1 state 2
Method: single particle / : Zhang H, Li Z, Li XZ

EMDB-37272: 
Cryo-EM structure of DSR2-DSAD1 state 1
Method: single particle / : Zhang H, Li Z, Li XZ
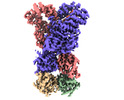
EMDB-37603: 
Cryo-EM structure of DSR2-DSAD1
Method: single particle / : Zhang H, Li Z, Li XZ

EMDB-38421: 
Cryo-EM structure of tail tube protein
Method: single particle / : Zhang H, Li Z, Li XZ

EMDB-17375: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17384: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17386: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17683: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17695: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17718: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p2v: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p36: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p3b: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pij: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8piz: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pjp: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-36870: 
Structure of full Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36871: 
In situ structure of RNA-dependent RNA polymerase in full BAV particles
Method: single particle / : Li Z, Cao S

EMDB-36872: 
Structure of VP9 in Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36880: 
Structure of partial Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36881: 
Structure of Banna virus core
Method: single particle / : Li Z, Cao S

EMDB-37378: 
Structure of full Banna virus
Method: single particle / : Li Z, Cao S

EMDB-37379: 
Structure of partial Banna virus
Method: single particle / : Li Z, Cao S

EMDB-37380: 
Structure of Banna virus core
Method: single particle / : Li Z, Cao S

PDB-8k43: 
In situ structure of RNA-dependent RNA polymerase in full BAV particles
Method: single particle / : Li Z, Cao S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search









 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
