+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Neisseria meningitidis Type IV pilus SB-GATDH variant | |||||||||
 Map data Map data | Raw map of the SB-GATDH variant of T4P | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Pilin / Pilin /  Extracellular / Extracellular /  Adhesion / Aggregation / PROTEIN FIBRIL Adhesion / Aggregation / PROTEIN FIBRIL | |||||||||
| Biological species |   Neisseria meningitidis 8013 (bacteria) Neisseria meningitidis 8013 (bacteria) | |||||||||
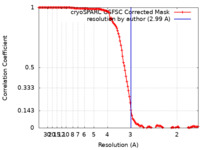
| Method | helical reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Fernandez-Martinez D / Dumenil G | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms. Authors: David Fernandez-Martinez / Youxin Kong / Sylvie Goussard / Agustin Zavala / Pauline Gastineau / Martial Rey / Gabriel Ayme / Julia Chamot-Rooke / Pierre Lafaye / Matthijn Vos / Ariel Mechaly ...Authors: David Fernandez-Martinez / Youxin Kong / Sylvie Goussard / Agustin Zavala / Pauline Gastineau / Martial Rey / Gabriel Ayme / Julia Chamot-Rooke / Pierre Lafaye / Matthijn Vos / Ariel Mechaly / Guillaume Duménil /  Abstract: Type IV pili (T4P) are prevalent, polymeric surface structures in pathogenic bacteria, making them ideal targets for effective vaccines. However, bacteria have evolved efficient strategies to evade ...Type IV pili (T4P) are prevalent, polymeric surface structures in pathogenic bacteria, making them ideal targets for effective vaccines. However, bacteria have evolved efficient strategies to evade type IV pili-directed antibody responses. Neisseria meningitidis are prototypical type IV pili-expressing Gram-negative bacteria responsible for life threatening sepsis and meningitis. This species has evolved several genetic strategies to modify the surface of its type IV pili, changing pilin subunit amino acid sequence, nature of glycosylation and phosphoforms, but how these modifications affect antibody binding at the structural level is still unknown. Here, to explore this question, we determine cryo-electron microscopy (cryo-EM) structures of pili of different sequence types with sufficiently high resolution to visualize posttranslational modifications. We then generate nanobodies directed against type IV pili which alter pilus function in vitro and in vivo. Cyro-EM in combination with molecular dynamics simulation of the nanobody-pilus complexes reveals how the different types of pili surface modifications alter nanobody binding. Our findings shed light on the impressive complementarity between the different strategies used by bacteria to avoid antibody binding. Importantly, we also show that structural information can be used to make informed modifications in nanobodies as countermeasures to these immune evasion mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17375.map.gz emd_17375.map.gz | 108.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17375-v30.xml emd-17375-v30.xml emd-17375.xml emd-17375.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17375_fsc.xml emd_17375_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17375.png emd_17375.png | 27.7 KB | ||
| Masks |  emd_17375_msk_1.map emd_17375_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17375.cif.gz emd-17375.cif.gz | 5.3 KB | ||
| Others |  emd_17375_additional_1.map.gz emd_17375_additional_1.map.gz emd_17375_half_map_1.map.gz emd_17375_half_map_1.map.gz emd_17375_half_map_2.map.gz emd_17375_half_map_2.map.gz | 191.3 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17375 http://ftp.pdbj.org/pub/emdb/structures/EMD-17375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17375 | HTTPS FTP |
-Related structure data
| Related structure data |  8p2vMC  8p36C  8p3bC  8pijC  8pizC  8pjpC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17375.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17375.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map of the SB-GATDH variant of T4P | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
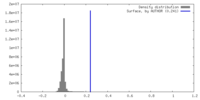
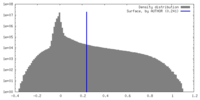
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17375_msk_1.map emd_17375_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
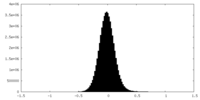
| Density Histograms |
-Additional map: DL sharpened map of the SB-GATDH variant of T4P
| File | emd_17375_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DL sharpened map of the SB-GATDH variant of T4P | ||||||||||||
| Projections & Slices |
| ||||||||||||
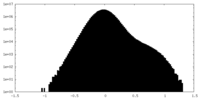
| Density Histograms |
-Half map: Half map A
| File | emd_17375_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
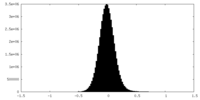
| Density Histograms |
-Half map: Half map B
| File | emd_17375_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
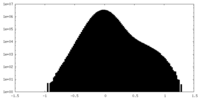
| Density Histograms |
- Sample components
Sample components
-Entire : PilE variant SB containing GATDH and G3P PTMs
| Entire | Name: PilE variant SB containing GATDH and G3P PTMs |
|---|---|
| Components |
|
-Supramolecule #1: PilE variant SB containing GATDH and G3P PTMs
| Supramolecule | Name: PilE variant SB containing GATDH and G3P PTMs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Neisseria meningitidis 8013 (bacteria) Neisseria meningitidis 8013 (bacteria) |
-Macromolecule #1: Neisseria meningitidis PilE variant SB-GATDH
| Macromolecule | Name: Neisseria meningitidis PilE variant SB-GATDH / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neisseria meningitidis 8013 (bacteria) Neisseria meningitidis 8013 (bacteria) |
| Molecular weight | Theoretical: 17.067182 KDa |
| Sequence | String: FTLIELMIVI AIVGILAAVA LPAYQDYTAR AQVSEAILLA EGQKSAVTEY YLNHGEWPGD NSSAGVATSA DIKGKYVQSV TVANGVITA QMASSNVNNE IKSKKLSLWA KRQNGSVKWF CGQPVTRTTA TATDVAAANG KTDDKINTKH LPSTCRDDSS A S |
-Macromolecule #2: SN-GLYCEROL-3-PHOSPHATE
| Macromolecule | Name: SN-GLYCEROL-3-PHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: G3P |
|---|---|
| Molecular weight | Theoretical: 172.074 Da |
| Chemical component information |  ChemComp-G3P: |
-Macromolecule #3: (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methy...
| Macromolecule | Name: (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide type: ligand / ID: 3 / Number of copies: 1 / Formula: WKE |
|---|---|
| Molecular weight | Theoretical: 292.286 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)