-Search query
-Search result
Showing 1 - 50 of 87 items for (author: arnold & p)

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
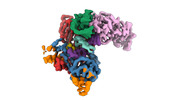
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
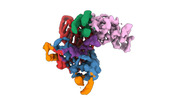
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-28615: 
MicroED structure of an Aeropyrum pernix protoglobin mutant
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

EMDB-28616: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

PDB-8eun: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

EMDB-26717: 
Gea2 closed/open conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

EMDB-26749: 
Consensus map of Gea2-Arf1 activation intermediate complex
Method: single particle / : Muccini A, Fromme JC

EMDB-26750: 
Focused map of DCB-HUS domains of the Gea2-Arf1 activation intermediate complex
Method: single particle / : Muccini A, Fromme JC

EMDB-26751: 
Focused map of GEF domain-Arf1 in the Gea2-Arf1 activation intermediate complex
Method: single particle / : Muccini A, Fromme JC

EMDB-26752: 
Focused map of HDS1,2,3 domains in the Gea2-Arf1 activation intermediate complex
Method: single particle / : Muccini A, Fromme JC

EMDB-26753: 
Focused map of the dimer interface of the Gea2-Arf1 activation intermediate complex
Method: single particle / : Muccini A, Fromme JC

EMDB-26754: 
Gea2 closed/closed conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

EMDB-26755: 
Consensus map of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26765: 
Focused map of the N-terminal region of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26766: 
Focused map of the GEF domain of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26769: 
Focused map of the C-terminal region of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26770: 
Gea2 open/open conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

EMDB-26771: 
Consensus map of Gea2 in open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26773: 
Focused map of the N-terminal region of Gea2 in the open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26774: 
Focused map of the GEF domain of Gea2 in the open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26775: 
Focused map of the C-terminal region of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26776: 
Focused map of the dimer interface of Gea2 in the open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26777: 
Focused map of the dimer interface of Gea2 in the closed conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26778: 
Focused map of the dimer interface of Gea2 in closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26779: 
Focused map of the closed state N-terminal region of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26780: 
Focused map of the closed state GEF domain of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26781: 
Focused map of the closed C-terminal region of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26783: 
Focused map of the open state N-terminal region of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26784: 
Focused map of the open state GEF domain of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26785: 
Focused map of the open state C-terminal region Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

EMDB-26797: 
Consensus map of Gea2 closed/open conformation
Method: single particle / : Muccini A, Fromme JC

PDB-7urr: 
Gea2 closed/open conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

PDB-7ut4: 
Gea2 closed/closed conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

PDB-7uth: 
Gea2 open/open conformation (composite structure)
Method: single particle / : Muccini A, Fromme JC

EMDB-25075: 
PR-RT portion of HIV-1 Pol
Method: single particle / : Lyumkis D, Passos D, Arnold E, Harrison J

EMDB-25074: 
Cryo-EM Structure of the RT component of the HIV-1 Pol Polyprotein
Method: single particle / : Lyumkis D, Passos D, Arnold E, Harrison JJE

EMDB-25165: 
Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein
Method: single particle / : Lyumkis D, Passos D

PDB-7sep: 
Cryo-EM Structure of the RT component of the HIV-1 Pol Polyprotein
Method: single particle / : Lyumkis D, Passos D, Arnold E, Harrison JJE

PDB-7sjx: 
Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein
Method: single particle / : Lyumkis D, Passos D, Arnold E, Harrison JJEK, Ruiz FX

EMDB-14457: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine
Method: single particle / : Singh AK, Das K

EMDB-14458: 
Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine
Method: single particle / : Singh AK, Das K

EMDB-14462: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with rilpivirine
Method: single particle / : Singh AK, Das K

EMDB-14463: 
Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with rilpivirine
Method: single particle / : Singh AK, Das K

EMDB-14465: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine
Method: single particle / : Singh AK, Das K

EMDB-14466: 
Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine
Method: single particle / : Singh AK, Das K

PDB-7z24: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine
Method: single particle / : Singh AK, Das K

PDB-7z29: 
Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine
Method: single particle / : Singh AK, Das K
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
