-Search query
-Search result
Showing 1 - 50 of 68 items for (author: anger & am)
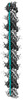
EMDB-19042: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19043: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19044: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (14pf) protofilament
Method: single particle / : Bangera M, Moores CA

PDB-8rc1: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-17645: 
Structure of a heteropolymeric type 4 pilus from a monoderm bacterium
Method: single particle / : Anger R, Pieulle L, Shahin M, Valette O, Le Guenno H, Kosta A, Pelicic V, Fronzes R

PDB-8pfb: 
Structure of a heteropolymeric type 4 pilus from a monoderm bacterium
Method: single particle / : Anger R, Pieulle L, Shahin M, Valette O, Le Guenno H, Kosta A, Pelicic V, Fronzes R

EMDB-14863: 
Bacteriophage T5 head - pb10-N-Ter fused to OVA
Method: single particle / : Schoehn G, Boulanger P, Benihoud K

EMDB-32033: 
14pf microtubule decorated with EML1-GFP
Method: helical / : Bangera M, Sirajuddin M

EMDB-12335: 
Structure of PSII-M
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-12336: 
Structure of PSII-I (PSII with Psb27, Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-12337: 
Structure of PSII-I prime (PSII with Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

PDB-7nho: 
Structure of PSII-M
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

PDB-7nhp: 
Structure of PSII-I (PSII with Psb27, Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

PDB-7nhq: 
Structure of PSII-I prime (PSII with Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-23211: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

EMDB-23215: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

EMDB-10711: 
Cryo-EM structure of a respiratory complex I F89A mutant
Method: single particle / : Parey K

PDB-6y79: 
Cryo-EM structure of a respiratory complex I F89A mutant
Method: single particle / : Parey K

EMDB-10891: 
Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Method: single particle / : Schulte L, Reitz J, Kudlinzki D, Hodirnau VV, Frangakis A, Schwalbe H

PDB-6ys3: 
Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Method: single particle / : Schulte L, Reitz J, Kudlinzki D, Hodirnau VV, Frangakis A, Schwalbe H

EMDB-10647: 
CryoEM structure of the wide type IV pilus (PilA4) from Thermus thermophilus
Method: helical / : Neuhaus A, Gold VAM

EMDB-10648: 
CryoEM structure of the narrow type IV pilus (PilA5) from Thermus thermophilus
Method: helical / : Neuhaus A, Gold VAM

PDB-6xxd: 
CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
Method: helical / : Neuhaus A, Gold VAM

PDB-6xxe: 
CryoEM structure of the type IV pilin PilA5 from Thermus thermophilus
Method: helical / : Neuhaus A, Gold VAM

EMDB-4531: 
Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide
Method: single particle / : Schulte L, Reitz J, Hodirnau VV, Kudlinzki D, Mao J, Glaubitz C, Frangakis A, Schwalbe H

PDB-6qdw: 
Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide
Method: single particle / : Schulte L, Reitz J, Hodirnau VV, Kudlinzki D, Mao J, Glaubitz C, Frangakis A, Schwalbe H

EMDB-10287: 
CLEM of resin-embedded GFP-Scs2 yeast cell shown in Figure 2A of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10299: 
CLEM of resin-embedded GFP-Ist2 yeast cell shown in Figure 2B of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10300: 
CLEM of resin-embedded Tcb3-GFP yeast cell shown in Figure 2C of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10301: 
CLEM of resin-embedded scs2/22 tcb1/2/3 deletion mutant yeast cell expressing GFP-Ist2 shown in Figure 2F of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10302: 
CLEM of resin-embedded scs2/22 ist2 deletion mutant yeast cell expressing Tcb3-GFP; shown in Figure 2G of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10303: 
CLEM of resin-embedded scs2/22 ist2 tcb1/2/3 deletion mutant yeast cell expressing Sec63-RFP from plasmid, shown in Figure 2H of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10304: 
CLEM of resin-embedded rtn1 yop1 deletion mutant yeast cell expressing Tcb3-GFP, shown in Figure 3C of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10306: 
CLEM of resin-embedded rtn1 yop1 deletion mutant yeast cell expressing Tcb3-GFP, shown in Figure 3E of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10308: 
cryo-ET of cryo-FIB milled yeast cell in which scs2/22 ist2 are deleted; shown in Figures 5A and S3B of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10309: 
cryo-ET of cryo-FIB milled yeast cell, in which scs2/22 ist2 are deleted, with high intracellular calcium; shown in Figures 5C and S3F
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-10310: 
cryo-ET of cryo-FIB milled yeast cell, in which Tcb3-GFP is overexpressed and scs2/22 ist2 tcb1/2 are deleted; shown in Figure 6 of publication
Method: electron tomography / : Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W

EMDB-4908: 
Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution
Method: single particle / : Safarian S, Hahn A, Kuehlbrandt W, Michel H

PDB-6rko: 
Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution
Method: single particle / : Safarian S, Hahn A, Kuehlbrandt W, Michel H

EMDB-20099: 
Prohead 2 of the phage T5
Method: single particle / : Huet A, Duda RL

EMDB-20122: 
non-decorated head of the phage T5
Method: single particle / : Huet A, Duda RL

EMDB-20123: 
decorated head of the phage T5
Method: single particle / : Huet A, Duda RL, Boulanger P, Conway JF

EMDB-20125: 
capsid of T5 virion
Method: single particle / : Huet A, Duda RL

PDB-6okb: 
Prohead 2 of the phage T5
Method: single particle / : Huet A, Duda RL, Boulanger P, Conway JF

PDB-6oma: 
non-decorated head of the phage T5
Method: single particle / : Huet A, Duda RL, Boulanger P, Conway JF

PDB-6omc: 
capsid of T5 virion
Method: single particle / : Huet A, Duda RL, Boulanger P, Conway JF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
