+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
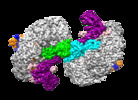
| Title | Structure of Csy-AcrIF24-dsDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Zhang L / Feng Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24. Authors: Lingguang Yang / Laixing Zhang / Peipei Yin / Hao Ding / Yu Xiao / Jianwei Zeng / Wenhe Wang / Huan Zhou / Qisheng Wang / Yi Zhang / Zeliang Chen / Maojun Yang / Yue Feng /  Abstract: CRISPR-Cas systems are prokaryotic adaptive immune systems and phages use anti-CRISPR proteins (Acrs) to counteract these systems. Here, we report the structures of AcrIF24 and its complex with the ...CRISPR-Cas systems are prokaryotic adaptive immune systems and phages use anti-CRISPR proteins (Acrs) to counteract these systems. Here, we report the structures of AcrIF24 and its complex with the crRNA-guided surveillance (Csy) complex. The HTH motif of AcrIF24 can bind the Acr promoter region and repress its transcription, suggesting its role as an Aca gene in self-regulation. AcrIF24 forms a homodimer and further induces dimerization of the Csy complex. Apart from blocking the hybridization of target DNA to the crRNA, AcrIF24 also induces the binding of non-sequence-specific dsDNA to the Csy complex, similar to AcrIF9, although this binding seems to play a minor role in AcrIF24 inhibitory capacity. Further structural and biochemical studies of the Csy-AcrIF24-dsDNA complexes and of AcrIF24 mutants reveal that the HTH motif of AcrIF24 and the PAM recognition loop of the Csy complex are structural elements essential for this non-specific dsDNA binding. Moreover, AcrIF24 and AcrIF9 display distinct characteristics in inducing non-specific DNA binding. Together, our findings highlight a multifunctional Acr and suggest potential wide distribution of Acr-induced non-specific DNA binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32440.map.gz emd_32440.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32440-v30.xml emd-32440-v30.xml emd-32440.xml emd-32440.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32440_fsc.xml emd_32440_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_32440.png emd_32440.png | 91.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32440 http://ftp.pdbj.org/pub/emdb/structures/EMD-32440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32440 | HTTPS FTP |
-Related structure data
| Related structure data |  7we6MC  7dtrC  7elmC  7elnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32440.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32440.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Csy-AcrIF24
| Entire | Name: Csy-AcrIF24 |
|---|---|
| Components |
|
-Supramolecule #1: Csy-AcrIF24
| Supramolecule | Name: Csy-AcrIF24 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: CRISPR type I-F/YPEST-associated protein Csy2
| Macromolecule | Name: CRISPR type I-F/YPEST-associated protein Csy2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 36.244074 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ ...String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ RRKNQRRLTR RLLPGFALVS REALLQQHLE TLRTTLPEAT TLDALLDLCR INFEPPATSS EEEASPPDAA WQ VRDKPGW LVPIPAGYNA LSPLYLPGEV RNARDRETPL RFVENLFGLG EWLSPHRVAA LSDLLWYHHA EPDKGLYRWS TPR FVEHAI A |
-Macromolecule #2: CRISPR-associated protein Csy3
| Macromolecule | Name: CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 37.623324 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNEQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG ...String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNEQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG EVARTWRFDA LAIGLRDFKA DAELDALAEL IASGLSGSGH VLLEVVAFAR IGDGQEVFPS QELILDKGDK KG QKSKTLY SVRDAAAIHS QKIGNALRTI DTWYPDEDGL GPIAVEPYGS VTSQGKAYRQ PKQKLDFYTL LDNWVLRDEA PAV EQQHYV IANLIRGGVF GEAEEK |
-Macromolecule #3: Type I-F CRISPR-associated endoribonuclease Cas6/Csy4
| Macromolecule | Name: Type I-F CRISPR-associated endoribonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 21.429477 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MDHYLDIRLR PDPEFPPAQL MSVLFGKLHQ ALVAQGGDRI GVSFPDLDES RSRLGERLRI HASADDLRAL LARPWLEGLR DHLQFGEPA VVPHPTPYRQ VSRVQAKSNP ERLRRRLMRR HDLSEEEARK RIPDTVARTL DLPFVTLRSQ STGQHFRLFI R HGPLQATA EEGGFTCYGL SKGGFVPWF |
-Macromolecule #4: AcrIF24
| Macromolecule | Name: AcrIF24 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 24.995271 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MNAIHIGPFS ITPAARGLHY GGLPHHQWTL YYGPREMAIK TLPDSYTSSE VRDEFSDIIA EFVIDARHRY APDVLELVNS DGDAVLARV AVSRLPEALS GCIPDDRFPY WLLTASRPRL GLPVTLNEYT ALAVELSAPP LAWITGLLPG EVLTHDAEEW R PPTSWELR ...String: MNAIHIGPFS ITPAARGLHY GGLPHHQWTL YYGPREMAIK TLPDSYTSSE VRDEFSDIIA EFVIDARHRY APDVLELVNS DGDAVLARV AVSRLPEALS GCIPDDRFPY WLLTASRPRL GLPVTLNEYT ALAVELSAPP LAWITGLLPG EVLTHDAEEW R PPTSWELR HVVGEGSFTG VSGAAAAALL GMSATNFRKY TAGDSAANRQ KISFAAWHYL LDRLGVKRAS |
-Macromolecule #6: Type I-F CRISPR-associated protein Csy1
| Macromolecule | Name: Type I-F CRISPR-associated protein Csy1 / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 49.313254 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLHPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EAVQALSDNA EQAREWRQAF I GITAVKGA ...String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLHPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EAVQALSDNA EQAREWRQAF I GITAVKGA PASHSLAKQL YFPLPGSGYH LLAPLFPTSL VHHVHALLRE ARFGDAAKAA REARSRQESW PHGFSEYPNL AI QKFGGTK PQNISQLNSE RYGENWLLPS LPPHWQRQDQ RAPIRHSSVF EHDFGRSPEV SRLTRTLQRL LAKTRHNNFT IRR YRAQLV GQICDEALQY AARLRELEPG WSATPGCQLH DAEQLWLDPL RAQTDETFLQ RRLRGDWPAE VGNRFANWLN RAVS SDSQI LGSPEAAQWS QELSKELTMF KEILEDERD |
-Macromolecule #5: RNA (60-MER)
| Macromolecule | Name: RNA (60-MER) / type: rna / ID: 5 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 19.265404 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCCG CGUCUACCUG GUUCACUGCC GUGUAGGCAG |
-Macromolecule #7: DNA (5'-D(P*GP*GP*AP*TP*GP*GP*CP*TP*TP*CP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*GP*GP*AP*TP*GP*GP*CP*TP*TP*CP*C)-3') / type: dna / ID: 7 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 16.574561 KDa |
| Sequence | String: (DA)(DG)(DC)(DA)(DG)(DC)(DT)(DG)(DC)(DA) (DC)(DC)(DT)(DT)(DC)(DA)(DC)(DG)(DG)(DC) (DG)(DG)(DG)(DC)(DT)(DT)(DG)(DA)(DT) (DG)(DT)(DC)(DC)(DG)(DC)(DG)(DT)(DC)(DT) (DA) (DC)(DC)(DT)(DG)(DG)(DA) ...String: (DA)(DG)(DC)(DA)(DG)(DC)(DT)(DG)(DC)(DA) (DC)(DC)(DT)(DT)(DC)(DA)(DC)(DG)(DG)(DC) (DG)(DG)(DG)(DC)(DT)(DT)(DG)(DA)(DT) (DG)(DT)(DC)(DC)(DG)(DC)(DG)(DT)(DC)(DT) (DA) (DC)(DC)(DT)(DG)(DG)(DA)(DT)(DG) (DG)(DC)(DT)(DT)(DC)(DC) |
-Macromolecule #8: DNA-r
| Macromolecule | Name: DNA-r / type: dna / ID: 8 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
| Molecular weight | Theoretical: 16.708691 KDa |
| Sequence | String: (DG)(DG)(DA)(DA)(DG)(DC)(DC)(DA)(DT)(DC) (DC)(DA)(DG)(DG)(DT)(DA)(DG)(DA)(DC)(DG) (DC)(DG)(DG)(DA)(DC)(DA)(DT)(DC)(DA) (DA)(DG)(DC)(DC)(DC)(DG)(DC)(DC)(DG)(DT) (DG) (DA)(DA)(DG)(DG)(DT)(DG) ...String: (DG)(DG)(DA)(DA)(DG)(DC)(DC)(DA)(DT)(DC) (DC)(DA)(DG)(DG)(DT)(DA)(DG)(DA)(DC)(DG) (DC)(DG)(DG)(DA)(DC)(DA)(DT)(DC)(DA) (DA)(DG)(DC)(DC)(DC)(DG)(DC)(DC)(DG)(DT) (DG) (DA)(DA)(DG)(DG)(DT)(DG)(DC)(DA) (DG)(DC)(DT)(DG)(DC)(DT) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DARK FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.3 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller