+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7eln | ||||||
|---|---|---|---|---|---|---|---|
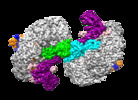
| Title | Structure of Csy-AcrIF24-dsDNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  IMMUNE SYSTEM/RNA/DNA / IMMUNE SYSTEM/RNA/DNA /  complex / complex /  inhibitor / inhibitor /  IMMUNE SYSTEM / IMMUNE SYSTEM /  IMMUNE SYSTEM-RNA-DNA complex IMMUNE SYSTEM-RNA-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationCRISPR-associated protein Csy1 / CRISPR-associated protein (Cas_Csy1) / CRISPR-associated protein Csy2 / CRISPR-associated protein (Cas_Csy2) / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) Similarity search - Domain/homology | ||||||
| Biological species |   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3 Å cryo EM / Resolution: 3 Å | ||||||
 Authors Authors | Zhang, L. / Feng, Y. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24. Authors: Lingguang Yang / Laixing Zhang / Peipei Yin / Hao Ding / Yu Xiao / Jianwei Zeng / Wenhe Wang / Huan Zhou / Qisheng Wang / Yi Zhang / Zeliang Chen / Maojun Yang / Yue Feng /  Abstract: CRISPR-Cas systems are prokaryotic adaptive immune systems and phages use anti-CRISPR proteins (Acrs) to counteract these systems. Here, we report the structures of AcrIF24 and its complex with the ...CRISPR-Cas systems are prokaryotic adaptive immune systems and phages use anti-CRISPR proteins (Acrs) to counteract these systems. Here, we report the structures of AcrIF24 and its complex with the crRNA-guided surveillance (Csy) complex. The HTH motif of AcrIF24 can bind the Acr promoter region and repress its transcription, suggesting its role as an Aca gene in self-regulation. AcrIF24 forms a homodimer and further induces dimerization of the Csy complex. Apart from blocking the hybridization of target DNA to the crRNA, AcrIF24 also induces the binding of non-sequence-specific dsDNA to the Csy complex, similar to AcrIF9, although this binding seems to play a minor role in AcrIF24 inhibitory capacity. Further structural and biochemical studies of the Csy-AcrIF24-dsDNA complexes and of AcrIF24 mutants reveal that the HTH motif of AcrIF24 and the PAM recognition loop of the Csy complex are structural elements essential for this non-specific dsDNA binding. Moreover, AcrIF24 and AcrIF9 display distinct characteristics in inducing non-specific DNA binding. Together, our findings highlight a multifunctional Acr and suggest potential wide distribution of Acr-induced non-specific DNA binding. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7eln.cif.gz 7eln.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7eln.ent.gz pdb7eln.ent.gz | 879.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7eln.json.gz 7eln.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/el/7eln https://data.pdbj.org/pub/pdb/validation_reports/el/7eln ftp://data.pdbj.org/pub/pdb/validation_reports/el/7eln ftp://data.pdbj.org/pub/pdb/validation_reports/el/7eln | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  31186MC  7dtrC  7elmC  7we6C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 5 types, 20 molecules BLCDEFGHMNOPQRISUVAX
| #1: Protein | Mass: 36244.074 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria)Gene: csy2, ALP65_00953, EQH76_13810, NCTC13437_01526, PACL_0128 Production host:   Escherichia coli (E. coli) / References: UniProt: B3G161 Escherichia coli (E. coli) / References: UniProt: B3G161#2: Protein | Mass: 37623.324 Da / Num. of mol.: 12 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) / Gene: IPC1505_30680, IPC36_28835 / Production host: Pseudomonas aeruginosa (bacteria) / Gene: IPC1505_30680, IPC36_28835 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A659BSG0 Escherichia coli (E. coli) / References: UniProt: A0A659BSG0#3: Protein | Mass: 21429.477 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) / Production host: Pseudomonas aeruginosa (bacteria) / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)#4: Protein | Mass: 24995.271 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) / Production host: Pseudomonas aeruginosa (bacteria) / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)#6: Protein | Mass: 49313.254 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) / Gene: csy1, ALP65_00954, IPC1505_30690 / Production host: Pseudomonas aeruginosa (bacteria) / Gene: csy1, ALP65_00954, IPC1505_30690 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A3A8DDU9 Escherichia coli (E. coli) / References: UniProt: A0A3A8DDU9 |
|---|
-RNA chain , 1 types, 2 molecules JT
| #5: RNA chain | Mass: 19265.404 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (bacteria) / Production host: Pseudomonas aeruginosa (bacteria) / Production host:   Escherichia coli (E. coli) / References: GenBank: 313291946 Escherichia coli (E. coli) / References: GenBank: 313291946 |
|---|
-DNA chain , 2 types, 4 molecules KYWZ
| #7: DNA chain | Mass: 16708.691 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria)#8: DNA chain | Mass: 16574.561 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
|---|
-Details
| Sequence details | The reference sequence database of chain I, S is WP_004343806.1 in GenBank. The reference sequence ...The reference sequence database of chain I, S is WP_004343806.1 in GenBank. The reference sequence database of chain U, V is WP_043084540.1 in GenBank. |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) | ||||||||||||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) | ||||||||||||||||||||||||
| Buffer solution | pH: 7 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: DARK FIELD |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.17.1_3660: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: RELION / Version: 3 / Category: particle selection | ||||||||||||||||||||||||
CTF correction | Type: NONE | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1564735 | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 364290 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj































































