+ Open data
Open data
- Basic information
Basic information
| Entry | 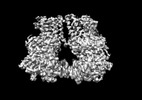 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hybrid aspen cellulose synthase-8 bound to UDP | |||||||||
 Map data Map data | autosharpened map local refinement in cryosparc | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information cellulose synthase (UDP-forming) / cellulose synthase (UDP-forming) /  cellulose synthase (UDP-forming) activity / cellulose biosynthetic process / cell wall organization / cellulose synthase (UDP-forming) activity / cellulose biosynthetic process / cell wall organization /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Populus tremula x Populus tremuloides (plant) Populus tremula x Populus tremuloides (plant) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Verma P / Zimmer J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8. Authors: Preeti Verma / Albert L Kwansa / Ruoya Ho / Yaroslava G Yingling / Jochen Zimmer /  Abstract: Cellulose is an abundant cell wall component of land plants. It is synthesized from UDP-activated glucose molecules by cellulose synthase, a membrane-integrated processive glycosyltransferase. ...Cellulose is an abundant cell wall component of land plants. It is synthesized from UDP-activated glucose molecules by cellulose synthase, a membrane-integrated processive glycosyltransferase. Cellulose synthase couples the elongation of the cellulose polymer with its translocation across the plasma membrane. Here, we present substrate and product-bound cryogenic electron microscopy structures of the homotrimeric cellulose synthase isoform-8 (CesA8) from hybrid aspen (poplar). UDP-glucose binds to a conserved catalytic pocket adjacent to the entrance to a transmembrane channel. The substrate's glucosyl unit is coordinated by conserved residues of the glycosyltransferase domain and amphipathic interface helices. Site-directed mutagenesis of a conserved gating loop capping the active site reveals its critical function for catalytic activity. Molecular dynamics simulations reveal prolonged interactions of the gating loop with the substrate molecule, particularly across its central conserved region. These transient interactions likely facilitate the proper positioning of the substrate molecule for glycosyl transfer and cellulose translocation. HIGHLIGHTS: Cryo-EM structures of substrate and product bound poplar cellulose synthase provide insights into substrate selectivitySite directed mutagenesis signifies a critical function of the ...HIGHLIGHTS: Cryo-EM structures of substrate and product bound poplar cellulose synthase provide insights into substrate selectivitySite directed mutagenesis signifies a critical function of the gating loop for catalysisMolecular dynamics simulations support persistent gating loop - substrate interactionsGating loop helps in positioning the substrate molecule to facilitate cellulose elongationConserved cellulose synthesis substrate binding mechanism across the kingdoms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29678.map.gz emd_29678.map.gz | 223.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29678-v30.xml emd-29678-v30.xml emd-29678.xml emd-29678.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29678.png emd_29678.png | 68.2 KB | ||
| Others |  emd_29678_half_map_1.map.gz emd_29678_half_map_1.map.gz emd_29678_half_map_2.map.gz emd_29678_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29678 http://ftp.pdbj.org/pub/emdb/structures/EMD-29678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29678 | HTTPS FTP |
-Related structure data
| Related structure data |  8g27MC  8g2jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29678.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29678.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | autosharpened map local refinement in cryosparc | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_29678_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
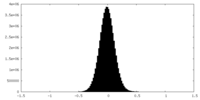
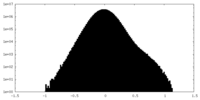
| Density Histograms |
-Half map: half map B
| File | emd_29678_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
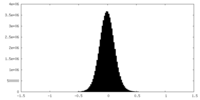
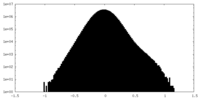
| Density Histograms |
- Sample components
Sample components
-Entire : Homotrimeric cellulose synthase complex
| Entire | Name: Homotrimeric cellulose synthase complex |
|---|---|
| Components |
|
-Supramolecule #1: Homotrimeric cellulose synthase complex
| Supramolecule | Name: Homotrimeric cellulose synthase complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Populus tremula x Populus tremuloides (plant) Populus tremula x Populus tremuloides (plant) |
-Macromolecule #1: Cellulose synthase
| Macromolecule | Name: Cellulose synthase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number:  cellulose synthase (UDP-forming) cellulose synthase (UDP-forming) |
|---|---|
| Source (natural) | Organism:   Populus tremula x Populus tremuloides (plant) Populus tremula x Populus tremuloides (plant) |
| Molecular weight | Theoretical: 112.483023 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHHHH HHHMMESGAP ICHTCGEQVG HDANGDLFVA CHECNYHICK SCFEYEIKEG RKVCLRCGSP YDENLLDDVE KKGSGNQST MASHLNNSQD VGIHARHISS VSTVDSEMND EYGNPIWKNR VESWKDKRNK KKKSNTKPET EPAQVPPEQQ M ENKPSAEA ...String: MHHHHHHHHH HHHMMESGAP ICHTCGEQVG HDANGDLFVA CHECNYHICK SCFEYEIKEG RKVCLRCGSP YDENLLDDVE KKGSGNQST MASHLNNSQD VGIHARHISS VSTVDSEMND EYGNPIWKNR VESWKDKRNK KKKSNTKPET EPAQVPPEQQ M ENKPSAEA SEPLSIVYPI PRNKLTPYRA VIIMRLIILG LFFHYRITNP VDSAFGLWLT SVICEIWFAF SWVLDQFPKW KP VNRETFI ERLSARYERE GEPSQLAAVD FFVSTVDPLK EPPLITANTV LSILAVDYPV DKVSCYVSDD GAAMLTFESL VET AEFARK WVPFCKKFSI EPRAPEFYFS QKIDYLKDKV QPSFVKERRA MKRDYEEYKV RVNALVAKAQ KTPDEGWTMQ DGTP WPGNN TRDHPGMIQV FLGNTGARDI EGNELPRLVY VSREKRPGYQ HHKKAGAENA LVRVSAVLTN APYILNLDCD HYVNN SKAV REAMCILMDP QVGRDVCYVQ FPQRFDGIDR SDRYANRNIV FFDVNMKGLD GIQGPMYVGT GCVFNRQALY GYGPPS MPR LRKGKESSSC FSCCCPTKKK PAQDPAEVYR DAKREDLNAA IFNLTEIDNY DDYERSMLIS QLSFEKTFGL SPVFIES TL MENGGVPESA NSSTLIKEAI HVIGCGFEEK TEWGKEIGWI YGSVTEDILS GFKMHCRGWR SIYCMPVRPA FKGSAPIN L SDRLHQVLRW ALGSVEIFFS RHCPFWYGYG GGRLKWLQRL AYINTIVYPF TSLPLIAYCT IPAVCLLTGK FIIPTLSNL ASMLFLGLFI SIIVTAVLEL RWSGVSIEDL WRNEQFWVIG GVSAHLFAVF QGFLKMLAGI DTNFTVTAKA ADDTEFGELY MVKWTTLLI PPTTLLIINI VGVVAGFSDA LNKGYEAWGP LFGKVFFAFW VILHLYPFLK GLMGRQNRTP TIVVLWSVLL T SVFSLVWV KINPFVNKVD NTLAGETCIS IDC |
-Macromolecule #3: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 0.8 µm / Nominal defocus min: 2.3000000000000003 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 15.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.8.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 188722 |
 Movie
Movie Controller
Controller



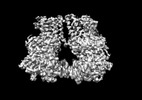



 Z
Z Y
Y X
X