[English] 日本語
 Yorodumi
Yorodumi- EMDB-27442: Cryo-EM structure of a HOPS core complex containing Vps33, Vps16,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 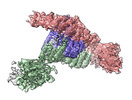 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | ||||||||||||
 Map data Map data | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  HOPS / membrane tethering / SNAREs / HOPS / membrane tethering / SNAREs /  membrane fusion / endolysosomal trafficking / membrane fusion / endolysosomal trafficking /  PROTEIN TRANSPORT PROTEIN TRANSPORT | ||||||||||||
| Biological species |   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.1 Å cryo EM / Resolution: 5.1 Å | ||||||||||||
 Authors Authors | Port SA / Farrell PD / Jeffrey PD / DiMaio F / Hughson FM | ||||||||||||
| Funding support |  Germany, Germany,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly Authors: Port SA / Farrell PD / Jeffrey PD / DiMaio F / Hughson FM | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27442.map.gz emd_27442.map.gz | 164.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27442-v30.xml emd-27442-v30.xml emd-27442.xml emd-27442.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27442_fsc.xml emd_27442_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27442.png emd_27442.png | 114.6 KB | ||
| Filedesc metadata |  emd-27442.cif.gz emd-27442.cif.gz | 7.7 KB | ||
| Others |  emd_27442_half_map_1.map.gz emd_27442_half_map_1.map.gz emd_27442_half_map_2.map.gz emd_27442_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27442 http://ftp.pdbj.org/pub/emdb/structures/EMD-27442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27442 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27442.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27442.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.426 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_27442_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_27442_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HOPS core complex
| Entire | Name: HOPS core complex |
|---|---|
| Components |
|
-Supramolecule #1: HOPS core complex
| Supramolecule | Name: HOPS core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Molecular weight | Theoretical: 171 KDa |
-Supramolecule #2: His-Vps33:Vps16
| Supramolecule | Name: His-Vps33:Vps16 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
-Supramolecule #3: Vps18
| Supramolecule | Name: Vps18 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
-Macromolecule #1: Vacuolar protein sorting-associated protein 33
| Macromolecule | Name: Vacuolar protein sorting-associated protein 33 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Molecular weight | Theoretical: 77.119617 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKHHHHHHHG AAGTSLYKKA GENLYFQGSM APRAGFDAEQ VRDKARKDLL HLLEGVRGKK NLVIEKDLAG PLGVIVKAST LRDYGVDNF FFLENKNTGT SQRNIVFIAR GESVRNAHAI AAQIKRIQRE SQTSHDFHIF WVPRRTLFSD KVLEEAGVLG D ANISELPL ...String: MKHHHHHHHG AAGTSLYKKA GENLYFQGSM APRAGFDAEQ VRDKARKDLL HLLEGVRGKK NLVIEKDLAG PLGVIVKAST LRDYGVDNF FFLENKNTGT SQRNIVFIAR GESVRNAHAI AAQIKRIQRE SQTSHDFHIF WVPRRTLFSD KVLEEAGVLG D ANISELPL YFFPLERDVL SLELNDSFRD LYLAKDPTPV FLLSRALMGI QKKHGLFPRI IGKGENAKRV ADLLSRMRQE LL AGEEAGE SDRAGLSPST TIESVIIIDR EVDFVTPLLT QLTYEGLIDE YFGIQNNQTD VDAVIVGAPA QSAASTSTAV PTN SSQSRK RKIQLDGSDS LYSQLRDANF AIVGSLLNTV ARRLKSDYES RHNTKTTAEL KEFVKKLPGY QAEQQSLKIH SNIA EEIIN YTRTEIFNKL LEVQQNLAAG ADPSSQFDSI EELVARDTPL PQVLRLLCLY SCISGGIKTK ELDHFRRLVL QGYGH QHLL TLHNLERLQM FLSKSSPLAS MITMSGSSGG PDQKTNYTYL RKQLRLIVDE VNEQDPNDIA YVYSGYAPLS IRLVQC VLQ KQYLLSITKG SGTVIAAGPV AGGGAQGWKG FEEIVKHARG PTFDEIQKGE DKAVKARALL SGSSGDKKTV FVVFVGG IT FTEIAALRFI AKQEEARRNI VICTTSIING NRMMNAAIET ATFEKTTVTT AAAQ |
-Macromolecule #2: Vacuolar protein sorting-associated protein 16
| Macromolecule | Name: Vacuolar protein sorting-associated protein 16 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Molecular weight | Theoretical: 94.204828 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MDTAHPTASW EQLGERFYRK IQLYTQVFDQ DFDLDNYIVT GAPYGGAIAL YRDDEKLVAY QPSRSSRPTI DICSLSGKLL RRISWDQGP IKGVGWSEDE KLLIVMVDGT VRCYFDLQSE FTQFSLGHGA EEHGVKSCRF YSHGLVALLG NNALVSVSSY D EPRPKLLA ...String: MDTAHPTASW EQLGERFYRK IQLYTQVFDQ DFDLDNYIVT GAPYGGAIAL YRDDEKLVAY QPSRSSRPTI DICSLSGKLL RRISWDQGP IKGVGWSEDE KLLIVMVDGT VRCYFDLQSE FTQFSLGHGA EEHGVKSCRF YSHGLVALLG NNALVSVSSY D EPRPKLLA SPPEGRVYSW NIIPPAYSLS RSVEVLLSVN QTIYVCDASE CEDRFLDIGP FSHIAVSPNG RFCALYTTTG KV HVITSDF QSRLSEHDTK SKIAPNYFEW CGNDAVVIAW DDEVHLVGPS GSLARFFYDS GRIHLIPDFD GVRILANDRC DFL QKVPDV IEEVFGLGAD SPASILLDAV EQLEMKSPKA DDNIQLIRPH LVEAVDTCVS AAGQEFSIHW QKQLLKAASF GKSV LDIYN SDDFVDMCET LRVLNAVRFY EVGLPLSYEQ YQRLSPSGLI SRLLNRHEYL LAIRIADHLR LPTDKIHVHW ASAKV RLGS EDDDTICRKI VEKLSGKPGI SFEVIARTAY EEGRTRLATE LLNHEPRAGR QVPLLLSMEE DELALDKAIE SGDTDL IYF VIHQLRRKLP LASFFRVVSS RPTASAMVEA LARNSDGDGN EDTALLKDLY YQDDRRLDGA SVFIREALQQ PETRTAS DK LDLAANLLQG NQKEHVFELG ALKEAKMLLR MQETFERDLT DSFVGLSVNQ TMFKLIKLGY HGRAKKIQSE FKVPERVA W WIRLQALVAK RDWNEIEEIS RQRKSPIGWE PFFNQVLQAG NPRLAATFIP KCTNLEPGQT ITMYEKCGMR VKAAEEAVR LKDTEAWNRL LEAAGRNTAE GREIERLGAT VFKK |
-Macromolecule #3: Vacuolar protein sorting-associated protein 18
| Macromolecule | Name: Vacuolar protein sorting-associated protein 18 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Molecular weight | Theoretical: 108.52757 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MALDLSSGFA AADAIANLQL ADATLPIFEV LPVQLQFSVA ADFVAGQAAN NVLVIALSNG RILRIDLNKP EDIDDIDLPK KPSEVGVIR RMFLDPTASH LIICTSLGEN YYLHSQSRQP RPLARLRGVV IESIAWSPAL PTQSTREILI GAADGNIYEA Y IETSTEFY ...String: MALDLSSGFA AADAIANLQL ADATLPIFEV LPVQLQFSVA ADFVAGQAAN NVLVIALSNG RILRIDLNKP EDIDDIDLPK KPSEVGVIR RMFLDPTASH LIICTSLGEN YYLHSQSRQP RPLARLRGVV IESIAWSPAL PTQSTREILI GAADGNIYEA Y IETSTEFY RREDKYLKLV QKLPDGPITG LWADSLPGHK DTRRVLVATS SRLFHWVGKI GRGHDSGGGA SIYDKLFEAE QP TVHALSG ASAAAMSMLV VSPDAEQPSP RFREDEVPER AFAWLSSHGV YHGKLLLKGP LSELGAKVFA EAKLLPRAQL ANP EGASRR QLSTEYVDAV ALTQWHIVSL VAGRVVIANR LTGSIIYDQT ILNPGQKAVG LCVDQQKSTF WLFTPQEIFE IVPR DEDRD IWKIMLQLQQ FDAALQYAHT PAEKDAVAIA SGDHLVSKGQ FLEAAAVYGK SSKPFEEVAL TFIDNEQPDA LRKYL LTKL GTYKKSAVMQ RVMIATWLIE VFMAKLNSLD DTIITGAELS ETLNPNQTKE QLEAVRAEFQ DFINKHKGDL DRKTVY DVI GSHGREEELL YYANAINDYN YVLSYWVQRE RWTEALKVLK KQTDPEVFYR YSSVLMTHAA TELVEILMRQ SNLNPRN LI PAMLEYDRNY KGPLAQNQAV RYLLYVVNQL GSTDSAVHNT LVSIYASHPS KDESALLEYL ESQGEEPNYD PDFALRLC I QHRRVLSCAH IYTSMGQYGA AVDLALAHDE VELASIIADR PISNPQLRKK LWLKVAKKVI SQQSDGIKTA IDFLRRCDL LKIEDLIPFF PDFVVIDDFK EEICAALEDY SRNIDALRRE MDEASQTAAN IKVDIAALDK RYAIVEPGEK CYACGLPLLS RQFFVFPCQ HAFHSDCLAR RVLEQAPPAK ARRIKECQVQ ISKGLVNGEK REAMIAELDA LIASACDYAI RRINEPFIKD D DDKDEWAL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: PELCO Easiglow, 10 mA/ 10 s discharge/ 30 s hold | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Blotting parameters: 30 seconds waiting time, blotting force 1, blot total 1. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.03 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm Bright-field microscopy / Cs: 0.03 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 3590 / Average exposure time: 5.6 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X


















