[English] 日本語
 Yorodumi
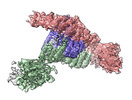
Yorodumi- PDB-8dit: Cryo-EM structure of a HOPS core complex containing Vps33, Vps16,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8dit | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords |  PROTEIN TRANSPORT / PROTEIN TRANSPORT /  HOPS / membrane tethering / SNAREs / HOPS / membrane tethering / SNAREs /  membrane fusion / endolysosomal trafficking membrane fusion / endolysosomal trafficking | ||||||||||||
| Biological species |   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | ||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.1 Å cryo EM / Resolution: 5.1 Å | ||||||||||||
 Authors Authors | Port, S.A. / Farrell, P.D. / Jeffrey, P.D. / DiMaio, F. / Hughson, F.M. | ||||||||||||
| Funding support |  Germany, Germany,  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly Authors: Port, S.A. / Farrell, P.D. / Jeffrey, P.D. / DiMaio, F. / Hughson, F.M. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8dit.cif.gz 8dit.cif.gz | 429.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8dit.ent.gz pdb8dit.ent.gz | 342 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8dit.json.gz 8dit.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/di/8dit https://data.pdbj.org/pub/pdb/validation_reports/di/8dit ftp://data.pdbj.org/pub/pdb/validation_reports/di/8dit ftp://data.pdbj.org/pub/pdb/validation_reports/di/8dit | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  Vacuole VacuoleMass: 77119.617 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Chaetomium thermophilum (fungus) / Plasmid: pQLinkH-Vps33:Vps16 / Details (production host): co-expression plasmid / Production host: Chaetomium thermophilum (fungus) / Plasmid: pQLinkH-Vps33:Vps16 / Details (production host): co-expression plasmid / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 codon+ Escherichia coli (E. coli) / Strain (production host): BL21 codon+ |
|---|---|
| #2: Protein |  Vacuole VacuoleMass: 94204.828 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Chaetomium thermophilum (fungus) / Plasmid: pQLinkH-Vps33:Vps16 / Details (production host): co-expression plasmid / Production host: Chaetomium thermophilum (fungus) / Plasmid: pQLinkH-Vps33:Vps16 / Details (production host): co-expression plasmid / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 codon+ Escherichia coli (E. coli) / Strain (production host): BL21 codon+ |
| #3: Protein |  Vacuole VacuoleMass: 108527.570 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Chaetomium thermophilum (fungus) / Plasmid: pQLinkN-Vps18 / Production host: Chaetomium thermophilum (fungus) / Plasmid: pQLinkN-Vps18 / Production host:   Escherichia coli (E. coli) / Variant (production host): C43 Escherichia coli (E. coli) / Variant (production host): C43 |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| ||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | ||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||
| Specimen | Conc.: 0.08 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||
| Specimen support | Details: PELCO Easiglow, 10 mA/ 10 s discharge/ 30 s hold / Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 | ||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 298 K Details: Blotting parameters: 30 seconds waiting time, blotting force 1, blot total 1 |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2000 nm / Nominal defocus min: 2000 nm / Cs Bright-field microscopy / Nominal defocus max: 2000 nm / Nominal defocus min: 2000 nm / Cs : 0.03 mm : 0.03 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 5.6 sec. / Electron dose: 50 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 3590 |
| EM imaging optics | Energyfilter name : GIF Bioquantum / Energyfilter slit width: 20 eV : GIF Bioquantum / Energyfilter slit width: 20 eV |
| Image scans | Width: 3838 / Height: 3710 / Movie frames/image: 32 / Used frames/image: 1-32 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 4092244 / Details: blob picker 60-280A (circular) | ||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 5.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 154917 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Refinement | Highest resolution: 5.1 Å |
 Movie
Movie Controller
Controller


 PDBj
PDBj