[English] 日本語
 Yorodumi
Yorodumi- EMDB-15796: Translating 70S ribosome in the unrotated state (A and P tRNAs) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Translating 70S ribosome in the unrotated state (A and P tRNAs) | |||||||||||||||
 Map data Map data | Post-processed (localfilter from cryoSPARC) map from Homogeneous Refinement | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | 70S /  bacterial / bacterial /  translation / translation /  high-resolution / high-resolution /  RIBOSOME RIBOSOME | |||||||||||||||
| Biological species |   Escherichia coli B (bacteria) Escherichia coli B (bacteria) | |||||||||||||||
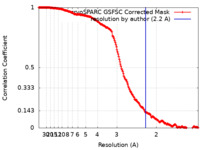
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.2 Å cryo EM / Resolution: 2.2 Å | |||||||||||||||
 Authors Authors | Fromm SA / O'Connor KM / Purdy M / Bhatt PR / Loughran G / Atkins JF / Jomaa A / Mattei S | |||||||||||||||
| Funding support |  Ireland, European Union, 4 items Ireland, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services. Authors: Simon A Fromm / Kate M O'Connor / Michael Purdy / Pramod R Bhatt / Gary Loughran / John F Atkins / Ahmad Jomaa / Simone Mattei /     Abstract: Our understanding of protein synthesis has been conceptualised around the structure and function of the bacterial ribosome. This complex macromolecular machine is the target of important ...Our understanding of protein synthesis has been conceptualised around the structure and function of the bacterial ribosome. This complex macromolecular machine is the target of important antimicrobial drugs, an integral line of defence against infectious diseases. Here, we describe how open access to cryo-electron microscopy facilities combined with bespoke user support enabled structural determination of the translating ribosome from Escherichia coli at 1.55 Å resolution. The obtained structures allow for direct determination of the rRNA sequence to identify ribosome polymorphism sites in the E. coli strain used in this study and enable interpretation of the ribosomal active and peripheral sites at unprecedented resolution. This includes scarcely populated chimeric hybrid states of the ribosome engaged in several tRNA translocation steps resolved at ~2 Å resolution. The current map not only improves our understanding of protein synthesis but also allows for more precise structure-based drug design of antibiotics to tackle rising bacterial resistance. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: The translating bacterial ribosome at 1.55 angstrom resolution by open access cryo-EM Authors: Fromm SA / O'Connor KM / Purdy M / Bhatt PR / Loughran G / Atkins JF / Jomaa A / Mattei S | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15796.map.gz emd_15796.map.gz | 64.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15796-v30.xml emd-15796-v30.xml emd-15796.xml emd-15796.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15796_fsc.xml emd_15796_fsc.xml | 18.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15796.png emd_15796.png | 102.8 KB | ||
| Masks |  emd_15796_msk_1.map emd_15796_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15796.cif.gz emd-15796.cif.gz | 5.1 KB | ||
| Others |  emd_15796_half_map_1.map.gz emd_15796_half_map_1.map.gz emd_15796_half_map_2.map.gz emd_15796_half_map_2.map.gz | 622.1 MB 622.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15796 http://ftp.pdbj.org/pub/emdb/structures/EMD-15796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15796 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15796.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15796.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed (localfilter from cryoSPARC) map from Homogeneous Refinement | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.731 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15796_msk_1.map emd_15796_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
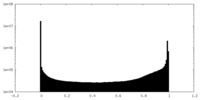
| Density Histograms |
-Half map: half map A
| File | emd_15796_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
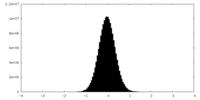
| Density Histograms |
-Half map: half map B
| File | emd_15796_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
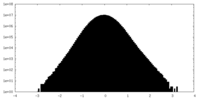
| Density Histograms |
- Sample components
Sample components
-Entire : 70S ribosome
| Entire | Name: 70S ribosome Ribosome Ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 70S ribosome
| Supramolecule | Name: 70S ribosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#53 |
|---|---|
| Source (natural) | Organism:   Escherichia coli B (bacteria) Escherichia coli B (bacteria) |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER / Details: Fischione 1070 | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 19449 / Average exposure time: 5.2 sec. / Average electron dose: 40.0 e/Å2 Details: saved in EER format; 16 exposures per hole/stage movement. 11 in outer ring around the hole edge, 5 in inner ring around the hole center. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X