+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13119 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. agalactiae BusR in complex with its busA-promotor DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmonoatomic cation transmembrane transporter activity / potassium ion transport / DNA-binding transcription factor activity Similarity search - Function | |||||||||
| Biological species |   Streptococcus agalactiae (bacteria) Streptococcus agalactiae (bacteria) | |||||||||
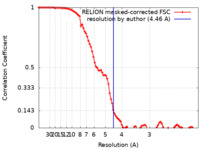
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.46 Å cryo EM / Resolution: 4.46 Å | |||||||||
 Authors Authors | Bandera AM / Witte G | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture. Authors: Adrian M Bandera / Joseph Bartho / Katja Lammens / David Jan Drexler / Jasmin Kleinschwärzer / Karl-Peter Hopfner / Gregor Witte /  Abstract: The cyclic dinucleotide second messenger c-di-AMP is a major player in regulation of potassium homeostasis and osmolyte transport in a variety of bacteria. Along with various direct interactions with ...The cyclic dinucleotide second messenger c-di-AMP is a major player in regulation of potassium homeostasis and osmolyte transport in a variety of bacteria. Along with various direct interactions with proteins such as potassium channels, the second messenger also specifically binds to transcription factors, thereby altering the processes in the cell on the transcriptional level. We here describe the structural and biochemical characterization of BusR from the human pathogen Streptococcus agalactiae. BusR is a member of a yet structurally uncharacterized subfamily of the GntR family of transcription factors that downregulates transcription of the genes for the BusA (OpuA) glycine-betaine transporter upon c-di-AMP binding. We report crystal structures of full-length BusR, its apo and c-di-AMP bound effector domain, as well as cryo-EM structures of BusR bound to its operator DNA. Our structural data, supported by biochemical and biophysical data, reveal that BusR utilizes a unique domain assembly with a tetrameric coiled-coil in between the binding platforms, serving as a molecular ruler to specifically recognize a 22 bp separated bipartite binding motif. Binding of c-di-AMP to BusR induces a shift in equilibrium from an inactivated towards an activated state that allows BusR to bind the target DNA, leading to transcriptional repression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13119.map.gz emd_13119.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13119-v30.xml emd-13119-v30.xml emd-13119.xml emd-13119.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13119_fsc.xml emd_13119_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13119.png emd_13119.png | 57.9 KB | ||
| Masks |  emd_13119_msk_1.map emd_13119_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_13119_half_map_1.map.gz emd_13119_half_map_1.map.gz emd_13119_half_map_2.map.gz emd_13119_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13119 http://ftp.pdbj.org/pub/emdb/structures/EMD-13119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13119 | HTTPS FTP |
-Related structure data
| Related structure data |  7oz3MC  7b5tC  7b5uC  7b5wC  7b5yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13119.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13119.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13119_msk_1.map emd_13119_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
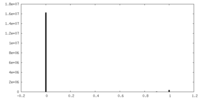
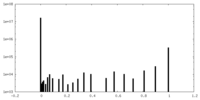
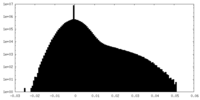
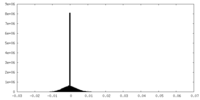
| Density Histograms |
-Half map: #2
| File | emd_13119_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13119_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
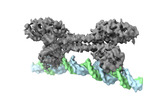
-Entire : transcriptional repressor BusR bound to target DNA
| Entire | Name: transcriptional repressor BusR bound to target DNA |
|---|---|
| Components |
|
-Supramolecule #1: transcriptional repressor BusR bound to target DNA
| Supramolecule | Name: transcriptional repressor BusR bound to target DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Streptococcus agalactiae (bacteria) Streptococcus agalactiae (bacteria) |
| Molecular weight | Theoretical: 122 KDa |
-Macromolecule #1: GntR family transcriptional regulator
| Macromolecule | Name: GntR family transcriptional regulator / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Streptococcus agalactiae (bacteria) Streptococcus agalactiae (bacteria) |
| Molecular weight | Theoretical: 23.88016 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: GPMVSEQSEI VTSKYQKIAV AVAQRIANGD YEVGEKLKSR TTIASTFNVS PETARKGLNI LADLQILTLK HGSGAIILSK EKAIEFLNQ YETSHSVAIL KGKIRDNIKA QQQEMEELAT LVDDFLLQTR AVSKQYPLAP YEIIVSEDSE HLGKSIGELN V WHQTGATI ...String: GPMVSEQSEI VTSKYQKIAV AVAQRIANGD YEVGEKLKSR TTIASTFNVS PETARKGLNI LADLQILTLK HGSGAIILSK EKAIEFLNQ YETSHSVAIL KGKIRDNIKA QQQEMEELAT LVDDFLLQTR AVSKQYPLAP YEIIVSEDSE HLGKSIGELN V WHQTGATI VAIEHEGKFI VSPGPFSVIE QGDHIFFVGD EDVYARMKTY FNLRMGL |
-Macromolecule #2: pBusA_rev
| Macromolecule | Name: pBusA_rev / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Streptococcus agalactiae (bacteria) Streptococcus agalactiae (bacteria) |
| Molecular weight | Theoretical: 46.903086 KDa |
| Sequence | String: (DA)(DT)(DA)(DG)(DG)(DA)(DT)(DC)(DC)(DG) (DT)(DG)(DT)(DC)(DT)(DT)(DA)(DT)(DC)(DT) (DA)(DT)(DA)(DA)(DA)(DG)(DT)(DG)(DA) (DC)(DG)(DT)(DT)(DG)(DA)(DG)(DT)(DT)(DA) (DT) (DC)(DG)(DT)(DA)(DA)(DA) ...String: (DA)(DT)(DA)(DG)(DG)(DA)(DT)(DC)(DC)(DG) (DT)(DG)(DT)(DC)(DT)(DT)(DA)(DT)(DC)(DT) (DA)(DT)(DA)(DA)(DA)(DG)(DT)(DG)(DA) (DC)(DG)(DT)(DT)(DG)(DA)(DG)(DT)(DT)(DA) (DT) (DC)(DG)(DT)(DA)(DA)(DA)(DA)(DG) (DG)(DG)(DT)(DA)(DG)(DT)(DC)(DA)(DC)(DT) (DT)(DT) (DT)(DT)(DT)(DA)(DC)(DT)(DC) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DT)(DA)(DA) (DA)(DC)(DT) (DG)(DC)(DC)(DG)(DA)(DG) (DA)(DC)(DA)(DG)(DA)(DC)(DC)(DA)(DT)(DA) (DA)(DC)(DT)(DA) (DT)(DA)(DA)(DT)(DA) (DT)(DC)(DA)(DT)(DG)(DT)(DA)(DT)(DG)(DG) (DT)(DC)(DT)(DC)(DT) (DT)(DT)(DA)(DT) (DG)(DT)(DC)(DA)(DA)(DC)(DA)(DC)(DC)(DT) (DT)(DG)(DC)(DT)(DT)(DA) (DG)(DA)(DG) (DA)(DA)(DG)(DC)(DT)(DT)(DT)(DA)(DT) |
-Macromolecule #3: pBusA_for
| Macromolecule | Name: pBusA_for / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Streptococcus agalactiae (bacteria) Streptococcus agalactiae (bacteria) |
| Molecular weight | Theoretical: 46.903062 KDa |
| Sequence | String: (DA)(DT)(DA)(DA)(DA)(DG)(DC)(DT)(DT)(DC) (DT)(DC)(DT)(DA)(DA)(DG)(DC)(DA)(DA)(DG) (DG)(DT)(DG)(DT)(DT)(DG)(DA)(DC)(DA) (DT)(DA)(DA)(DA)(DG)(DA)(DG)(DA)(DC)(DC) (DA) (DT)(DA)(DC)(DA)(DT)(DG) ...String: (DA)(DT)(DA)(DA)(DA)(DG)(DC)(DT)(DT)(DC) (DT)(DC)(DT)(DA)(DA)(DG)(DC)(DA)(DA)(DG) (DG)(DT)(DG)(DT)(DT)(DG)(DA)(DC)(DA) (DT)(DA)(DA)(DA)(DG)(DA)(DG)(DA)(DC)(DC) (DA) (DT)(DA)(DC)(DA)(DT)(DG)(DA)(DT) (DA)(DT)(DT)(DA)(DT)(DA)(DG)(DT)(DT)(DA) (DT)(DG) (DG)(DT)(DC)(DT)(DG)(DT)(DC) (DT)(DC)(DG)(DG)(DC)(DA)(DG)(DT)(DT)(DT) (DA)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DG) (DA)(DG)(DT)(DA)(DA)(DA)(DA)(DA)(DA)(DG) (DT)(DG)(DA)(DC) (DT)(DA)(DC)(DC)(DC) (DT)(DT)(DT)(DT)(DA)(DC)(DG)(DA)(DT)(DA) (DA)(DC)(DT)(DC)(DA) (DA)(DC)(DG)(DT) (DC)(DA)(DC)(DT)(DT)(DT)(DA)(DT)(DA)(DG) (DA)(DT)(DA)(DA)(DG)(DA) (DC)(DA)(DC) (DG)(DG)(DA)(DT)(DC)(DC)(DT)(DA)(DT) |
-Macromolecule #4: (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-...
| Macromolecule | Name: (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide type: ligand / ID: 4 / Number of copies: 2 / Formula: 2BA |
|---|---|
| Molecular weight | Theoretical: 658.412 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
Details: degassed, filtered | |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K Details: 0.05% beta-octyl glycoside added prior to plunge freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 45.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | RCK_C domains were build by rigid body fit of the model 7B5U. The flexible linker between RCK_C domain and coiled-coil domain were build de novo. | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 177 | ||||||
| Output model |  PDB-7oz3: |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X