3NHK
 
 | |
1VEQ
 
 | | Mycobacterium smegmatis Dps Hexagonal form | | Descriptor: | FE (III) ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-03 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
4A6P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
8BW8
 
 | |
8BW9
 
 | | Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP | | Descriptor: | Connector enhancer of KSR protein CNK, Dual specificity mitogen-activated protein kinase kinase dSOR1, KSR, ... | | Authors: | Maisonneuve, P, Fronzes, R, Sicheri, F. | | Deposit date: | 2022-12-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The CNK-HYP scaffolding complex promotes RAF activation by enhancing KSR-MEK interaction.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1VJZ
 
 | |
4GZT
 
 | | N2 neuraminidase D151G mutant of A/Tanzania/205/2010 H3N2 in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
1VKN
 
 | |
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
1H1N
 
 | |
4HGP
 
 | |
6EXC
 
 | |
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
1ADG
 
 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-SELENAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
3MZS
 
 | | Crystal Structure of Cytochrome P450 CYP11A1 in complex with 22-hydroxy-cholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Cholesterol side-chain cleavage enzyme, ISOPROPYL ALCOHOL, ... | | Authors: | Stout, C.D, Annalora, A, Mast, N, Pikuleva, I. | | Deposit date: | 2010-05-12 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Three-step Sequential Catalysis by the Cholesterol Side Chain Cleavage Enzyme CYP11A1.
J.Biol.Chem., 286, 2011
|
|
1H4P
 
 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2001-05-11 | | Release date: | 2003-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1A4Z
 
 | |
3ATO
 
 | | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Kumasaka, T. | | Deposit date: | 2011-01-06 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation
To be Published
|
|
4H34
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation
To be Published
|
|
1GZ3
 
 | | Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-14 | | Release date: | 2003-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
7XGW
 
 | |
4ACZ
 
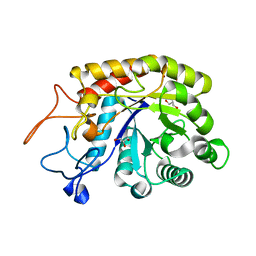 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1VQ9
 
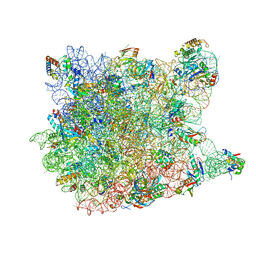 | |
1VM6
 
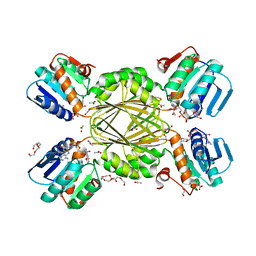 | |
1H4H
 
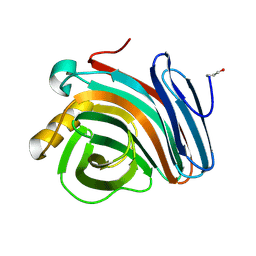 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
