5A3W
 
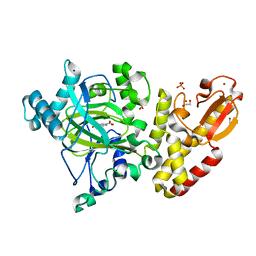 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
2W4H
 
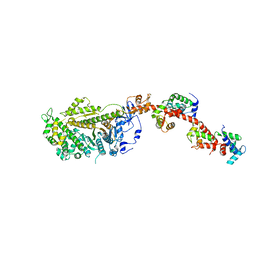 | | Isometrically contracting insect asynchronous flight muscle quick frozen after a quick release step | | Descriptor: | MYOSIN HEAVY CHAIN, SKELETAL MUSCLE, ADULT, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-11-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Changes in Isometrically Contracting Insect Flight Muscle Trapped Following a Mechanical Perturbation.
Plos One, 7, 2012
|
|
6W00
 
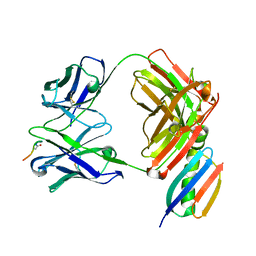 | | Crystal structure of Fab239 in complex with NPNA2 peptide from circumsporozoite protein | | Descriptor: | Fab239 heavy chain, Fab239 light chain, Immunoglobulin G-binding protein G, ... | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
4H9U
 
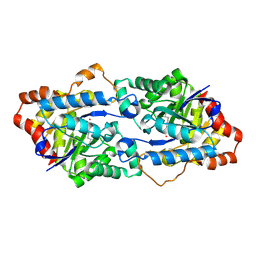 | | Structure of Geobacillus kaustophilus lactonase, wild-type with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
5A5I
 
 | | Cytochrome 2C9 P450 inhibitor complex | | Descriptor: | CYTOCHROME P450 2C9, N-[4-(3-chloranyl-4-cyano-phenoxy)cyclohexyl]-1,1,1-tris(fluoranyl)methanesulfonamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Skerratt, S.E, de Groot, M.J, Phillips, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel Binding Pocket for Cyp 2C9 Inhibitors: Crystallography, Pharmacophore Modelling and Inhibitor Sar.
Med.Chem.Comm., 7, 2016
|
|
4GV7
 
 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor ME0328 | | Descriptor: | 2-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
2WBC
 
 | | REFINED CRYSTAL STRUCTURE (2.3 ANGSTROM) OF A WINGED BEAN CHYMOTRYPSIN INHIBITOR AND LOCATION OF ITS SECOND REACTIVE SITE | | Descriptor: | CHYMOTRYPSIN INHIBITOR | | Authors: | Dattagupta, J.K, Podder, A, Chakrabarti, C, Sen, U, Mukhopadhyay, D, Dutta, S.K, Singh, M. | | Deposit date: | 1997-11-26 | | Release date: | 1998-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structure (2.3 A) of a double-headed winged bean alpha-chymotrypsin inhibitor and location of its second reactive site.
Proteins, 35, 1999
|
|
1J2E
 
 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
4H56
 
 | | Crystal structure of the Clostridium perfringens NetB toxin in the membrane inserted form | | Descriptor: | Necrotic enteritis toxin B | | Authors: | Savva, C.G, Fernandes da Costa, S.P, Bokori-Brown, M, Naylor, C, Cole, A.R, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture and Functional Analysis of NetB, a Pore-forming Toxin from Clostridium perfringens.
J.Biol.Chem., 288, 2013
|
|
5ZLP
 
 | | Crystal structure of glutamine synthetase from helicobacter pylori | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Analysis of Glutamine Synthetase from Helicobacter pylori.
Sci Rep, 8, 2018
|
|
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | Descriptor: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | Authors: | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | Deposit date: | 2005-05-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
5AGM
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-oxoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-OXOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | OPTINEURIN, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
4XI9
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RBL2) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Retinoblastoma-like protein 2, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2VUS
 
 | | Crystal structure of unliganded NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
1JPW
 
 | | Crystal Structure of a Human Tcf-4 / beta-Catenin Complex | | Descriptor: | BETA-CATENIN, transcription factor 7-like 2 | | Authors: | Poy, F, Lepourcelet, M, Shivdasani, R.A, Eck, M.J. | | Deposit date: | 2001-08-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a human Tcf4-beta-catenin complex.
Nat.Struct.Biol., 8, 2001
|
|
4XBM
 
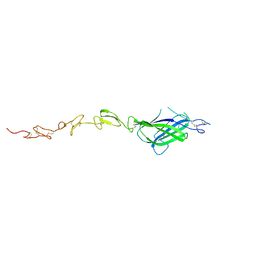 | | X-ray crystal structure of Notch ligand Delta-like 1 | | Descriptor: | Delta-like protein 1, alpha-L-fucopyranose | | Authors: | Kershaw, N.J, Burgess, A.W, Church, N.L, Luo, C.S, Adam, T.E. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Notch ligand delta-like1: X-ray crystal structure and binding affinity.
Biochem.J., 468, 2015
|
|
4H7M
 
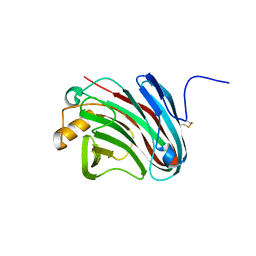 | |
1K2U
 
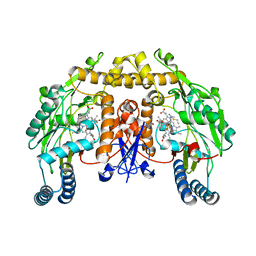 | | Structure of rat brain nNOS heme domain complexed with S-ethyl-N-[4-(trifluoromethyl)phenyl] isothiourea | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
2CWR
 
 | |
2VOX
 
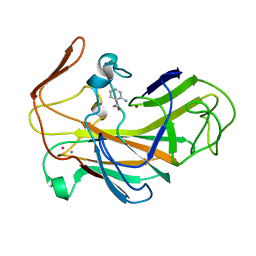 | | An oxidized tryptophan facilitates copper-binding in Methylococcus capsulatus secreted protein MopE. The structure of mercury soaked MopE to 1.9AA | | Descriptor: | CALCIUM ION, COPPER (II) ION, MERCURY (II) ION, ... | | Authors: | Helland, R, Fjellbirkeland, A, Karlsen, O.A, Ve, T, Lillehaug, J.R, Jensen, H.B. | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-18 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Oxidized Tryptophan Facilitates Copper Binding in Methylococcus Capsulatus-Secreted Protein Mope.
J.Biol.Chem., 283, 2008
|
|
6WBQ
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Tubastatin A | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-methyl-3,4-dihydro-1~{H}-pyrido[4,3-b]indol-5-yl)methyl]-~{N}-oxidanyl-benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
6VZG
 
 | | Cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Leschziner, A.E, Baker, R.W. | | Deposit date: | 2020-02-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into assembly and function of the RSC chromatin remodeling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6VUI
 
 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
2VOT
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (5R,6R,7S,8R)-6,7,8-trihydroxy-5-(hydroxymethyl)-2-[(phenylamino)methyl]-5,6,7,8-tetrahydro-1H-imidazo[1,2-a]pyridin-4-ium, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.N, Offen, W.A, Smith, N, Dumon, C, Moreland, C, Gratien, J, Heck, M.-P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-20 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
