1UBC
 
 | | Structure of Reca Protein | | Descriptor: | RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
6UFM
 
 | |
2ZYP
 
 | | X-ray structure of hen egg-white lysozyme with poly(allyl amine) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Ito, L, Tomita, S, Yamaguchi, H, Shiraki, K. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | X-ray structure of hen egg-white lysozyme with poly(allyl amine)
to be published
|
|
2ZZC
 
 | | Crystal structure of NADP(H):human thioredoxin reductase I | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Lo, Y.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2009-02-09 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Terpyridine-platinum(II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase 1.
J.Inorg.Biochem., 103, 2009
|
|
4I5D
 
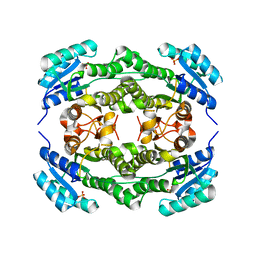 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in its apo form | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase, SULFATE ION | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
3WRJ
 
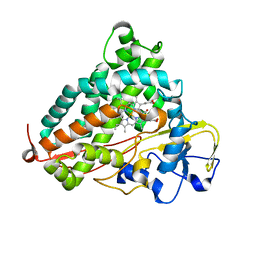 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3EZF
 
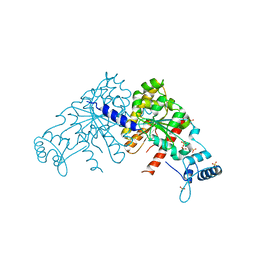 | | Partition Protein | | Descriptor: | ParA, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
6UH7
 
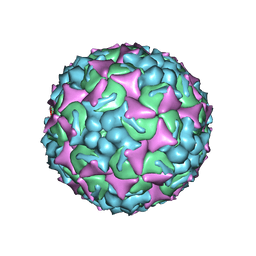 | | EV-A71 strain 11316 complexed with MADAL compound 30 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
6U4Y
 
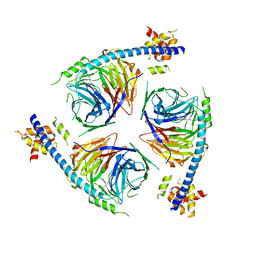 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1QRV
 
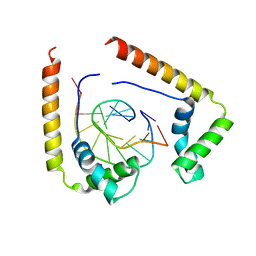 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HMG-D AND DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3'), HIGH MOBILITY GROUP PROTEIN D, SODIUM ION | | Authors: | Murphy IV, F.V, Sweet, R.M, Churchill, M.E.A. | | Deposit date: | 1999-06-15 | | Release date: | 1999-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition.
EMBO J., 18, 1999
|
|
1BW9
 
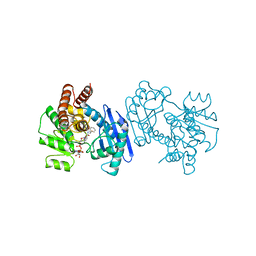 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1GBY
 
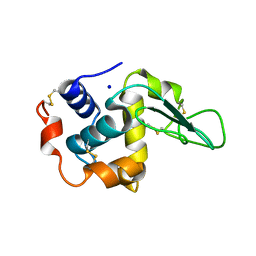 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
3WUA
 
 | |
3WW6
 
 | | Crystal Structure of hen egg white lysozyme mutant N46D/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
3NV5
 
 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
1U13
 
 | |
1GDX
 
 | |
6E18
 
 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
1GEM
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450CAM, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
3WX7
 
 | | Crystal structure of COD | | Descriptor: | ACETATE ION, CALCIUM ION, Chitin oligosaccharide deacetylase, ... | | Authors: | Park, S.-Y, Sugiyama, K, Hirano, T, Nishio, T. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus.
FEBS Lett., 589, 2015
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
1U4O
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedicarboxylic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,6-DICARBOXYNAPHTHALENE, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U5C
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1GEZ
 
 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
1GF4
 
 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
