1I77
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ESSEX 6 | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Einsle, O, Foerster, S, Mann, K.H, Fritz, G, Messerschmidt, A, Kroneck, P.M.H. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spectroscopic investigation and determination of reactivity and structure of the tetraheme cytochrome c3 from Desulfovibrio desulfuricans Essex 6.
Eur.J.Biochem., 268, 2001
|
|
1DIY
 
 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G, Ginell, S.L, Smith, W.L, Garavito, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The productive conformation of arachidonic acid bound to prostaglandin synthase.
Science, 289, 2000
|
|
1I4K
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1I5H
 
 | |
1I7W
 
 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I8H
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH HUMAN TAU PHOSPHOTHREONINE PEPTIDE | | Descriptor: | MICROTUBULE-ASSOCIATED PROTEIN TAU, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B33
 
 | | STRUCTURE OF LIGHT HARVESTING COMPLEX OF ALLOPHYCOCYANIN ALPHA AND BETA CHAINS/CORE-LINKER COMPLEX AP*LC7.8 | | Descriptor: | ALLOPHYCOCYANIN, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Reuter, W, Wiegand, G, Huber, R, Than, M.E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis at 2.2 A of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1I6C
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-02 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1IAK
 
 | |
1HVQ
 
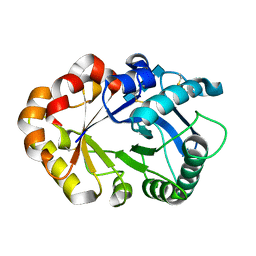 | | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEVAMINE A | | Authors: | Terwisscha Van Scheltinga, A.C, Kalk, K.H, Beintema, J.J, Dijkstra, B.W. | | Deposit date: | 1994-10-13 | | Release date: | 1995-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor.
Structure, 2, 1994
|
|
1BCD
 
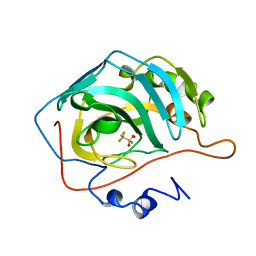 | |
1HXV
 
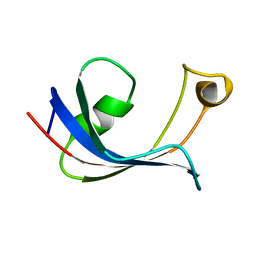 | | PPIASE DOMAIN OF THE MYCOPLASMA GENITALIUM TRIGGER FACTOR | | Descriptor: | TRIGGER FACTOR | | Authors: | Vogtherr, M, Parac, T.N, Maurer, M, Pahl, A, Fiebig, K. | | Deposit date: | 2001-01-17 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and dynamics of the peptidyl-prolyl cis-trans isomerase domain of the trigger factor from Mycoplasma genitalium compared to FK506-binding protein.
J.Mol.Biol., 318, 2002
|
|
1AO8
 
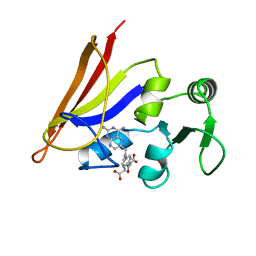 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE, NMR, 21 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Gargaro, A.R, Soteriou, A, Frenkiel, T.A, Bauer, C.J, Birdsall, B, Polshakov, V.I, Barsukov, I.L, Roberts, G.C.K, Feeney, J. | | Deposit date: | 1997-07-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate.
J.Mol.Biol., 277, 1998
|
|
1APF
 
 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
1HHV
 
 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
1HKJ
 
 | | Crystal structure of human chitinase in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1HPC
 
 | | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | | Descriptor: | 5-[(3S)-1,2-dithiolan-3-yl]pentanoic acid, H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM, LIPOIC ACID | | Authors: | Pares, S, Cohen-Addad, C, Sieker, L, Neuburger, M, Douce, R. | | Deposit date: | 1994-02-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1AVQ
 
 | |
1B75
 
 | |
1HTL
 
 | |
1HMV
 
 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
1HKK
 
 | | High resoultion crystal structure of human chitinase in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE-1, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase.
J.Biol.Chem., 278, 2003
|
|
1HN3
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL 37 AMINO ACIDS OF THE MOUSE ARF TUMOR SUPPRESSOR PROTEIN | | Descriptor: | P19 ARF PROTEIN | | Authors: | DiGiammarino, E.L, Filippov, I, Weber, J.D, Bothner, B, Kriwacki, R.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the p53 regulatory domain of the p19Arf tumor suppressor protein.
Biochemistry, 40, 2001
|
|
