1NCS
 
 | | NMR STUDY OF SWI5 ZINC FINGER DOMAIN 1 | | Descriptor: | TRANSCRIPTIONAL FACTOR SWI5, ZINC ION | | Authors: | Dutnall, R.N, Neuhaus, D, Rhodes, D. | | Deposit date: | 1996-02-26 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first zinc finger domain of SWI5: a novel structural extension to a common fold.
Structure, 4, 1996
|
|
4P2B
 
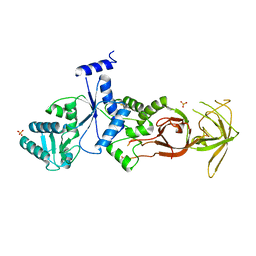 | |
1N7G
 
 | | Crystal Structure of the GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP-rhamnose. | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE-RHAMNOSE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the MUR1 GDP-mannose 4,6-dehydratase
from A. thaliana: Implications for ligand binding and
specificity.
Biochemistry, 41, 2002
|
|
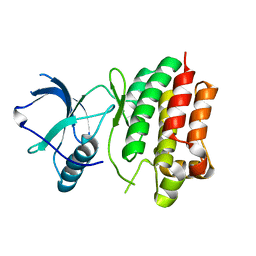
4P2K
 
 | |
1N83
 
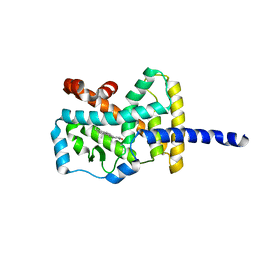 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
4P3L
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CHROMOHALOBACTER SALEXIGENS DSM 3043 (Csal_2479), TARGET EFI-510085, WITH BOUND GLUCURONATE, SPG P6122 | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-09 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1NKD
 
 | |
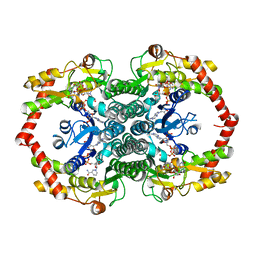
4P2X
 
 | | Swapped Dimer of Mycobacterial Adenylyl cyclase Rv1625c: Form 2 | | Descriptor: | Adenylate cyclase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Barathy, D.V, Mattoo, R, Visweswariah, S.S, Suguna, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New structural forms of a mycobacterial adenylyl cyclase Rv1625c.
Iucrj, 1, 2014
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
4P3V
 
 | |
1NKM
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1N8V
 
 | | Chemosensory Protein in complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4P32
 
 | | Crystal structure of E. coli LptB in complex with ADP-magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P3Z
 
 | | Chlamydia pneumoniae CopN (D29 construct) | | Descriptor: | CopN, GLYCEROL, SULFATE ION | | Authors: | Nawrotek, A, Guimaraes, B.G, Knossow, M, Gigant, B. | | Deposit date: | 2014-03-10 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
J.Biol.Chem., 289, 2014
|
|
1NKV
 
 | | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13 | | Descriptor: | HYPOTHETICAL PROTEIN yjhP | | Authors: | Kuzin, A, Manor, P, Benach, J, Smith, P, Rost, B, Xiao, R, Montelione, G, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13
To be published
|
|
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
1N90
 
 | |
4P43
 
 | |
1NL2
 
 | | BOVINE PROTHROMBIN FRAGMENT 1 IN COMPLEX WITH CALCIUM AND LYSOPHOSPHOTIDYLSERINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, M, Huang, G, Furie, B, Seaton, B, Furie, B.C. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins.
Nat.Struct.Biol., 10, 2003
|
|
4P48
 
 | | The structure of a chicken anti-cardiac Troponin I scFv | | Descriptor: | Antibody scFv 180 | | Authors: | Conroy, P.J, Law, R.H.P, Gillgunn, S, Hearty, S, Caradoc-Davies, T.T, Llyod, G, O'Kennedy, R.J, Whisstock, J.C. | | Deposit date: | 2014-03-12 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Reconciling the structural attributes of avian antibodies.
J.Biol.Chem., 289, 2014
|
|
1N9E
 
 | | Crystal structure of Pichia pastoris Lysyl Oxidase PPLO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2002-11-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Pichia pastoris Lysyl Oxidase
Biochemistry, 42, 2003
|
|
4P4B
 
 | |
1NLA
 
 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
4P4D
 
 | |
1NAP
 
 | |
