2CO7
 
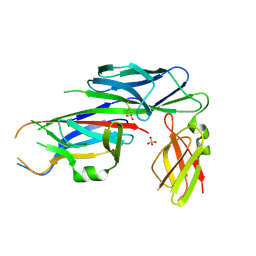 | | Salmonella enterica SafA pilin in complex with the SafB chaperone (Type II) | | Descriptor: | PUTATIVE FIMBRIAE ASSEMBLY CHAPERONE, SAFA PILUS SUBUNIT, SULFATE ION | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
7ZA7
 
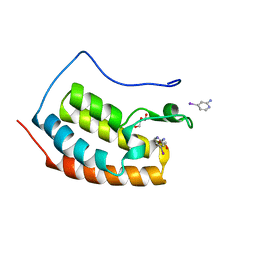 | | BRD4 in complex with FragLite5 | | Descriptor: | 4-iodanylpyridin-2-amine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5T5F
 
 | |
2CJJ
 
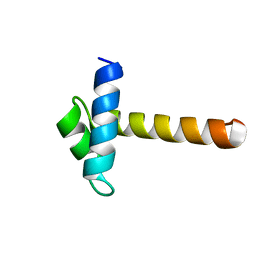 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
7ZA9
 
 | | BRD4 in complex with FragLite7 | | Descriptor: | 4-iodanyl-3~{H}-pyridin-2-one, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZEN
 
 | | BRD4 in complex with FragLite23 | | Descriptor: | 2-(4-bromo-1H-pyrazol-1-yl)ethan-1-ol, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5M84
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanyl-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog
To Be Published
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
5MBC
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
6Z4E
 
 | | The structure of the C-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
2PHY
 
 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Borgstahl, G.E.O, Getzoff, E.D. | | Deposit date: | 1995-04-12 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore.
Biochemistry, 34, 1995
|
|
6GJW
 
 | | Structure of XIAP-BIR1 domain in complex with an NF023 analog | | Descriptor: | 4-[[3-[[3-[(4,8-disulfonatonaphthalen-1-yl)carbamoyl]phenyl]carbamoylamino]phenyl]carbonylamino]naphthalene-1,5-disulfonate, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Sorrentino, L, Cossu, F, Malkoc, B, Zaffaroni, M, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
6R7G
 
 | | Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family | | Descriptor: | Coat protein, polyU RNA | | Authors: | Grinzato, A, Kandiah, E, Lico, C, Betti, C, Baschieri, S, Zanotti, G. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family.
Nat.Chem.Biol., 16, 2020
|
|
2H7B
 
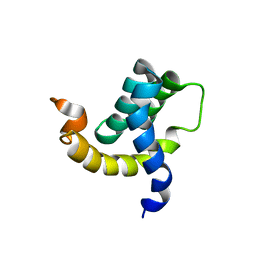 | | Solution structure of the eTAFH domain from the human leukemia-associated fusion protein AML1-ETO | | Descriptor: | Core-binding factor, ML1-ETO | | Authors: | Plevin, M.J, Zhang, J, Guo, C, Roeder, R.G, Ikura, M. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2PPA
 
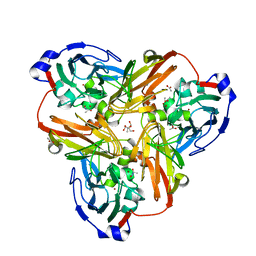 | |
6GKR
 
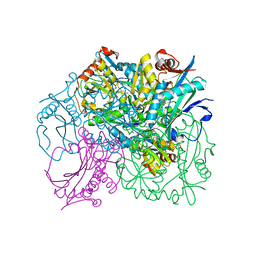 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PLP-form (holo-form) | | Descriptor: | ACETATE ION, Branched-chain-amino-acid aminotransferase, CHLORIDE ION, ... | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6R7Z
 
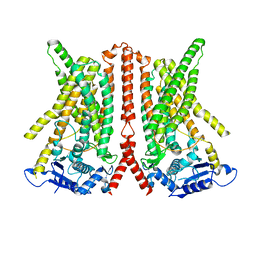 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4BPA
 
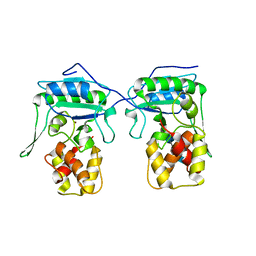 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
6GQ1
 
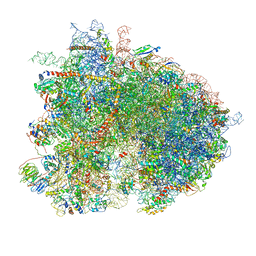 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Demeshkina, N, Mancera-Martinez, E, Melnikov, S, Simonetti, A, Myasnikov, A, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
6GUP
 
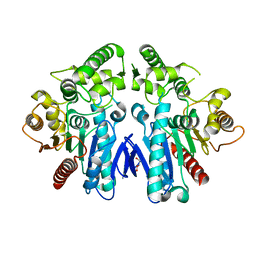 | | Siderophore hydrolase EstB from Aspergillus fumigatus | | Descriptor: | GLYCEROL, Siderophore biosynthesis lipase/esterase, putative | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5IT9
 
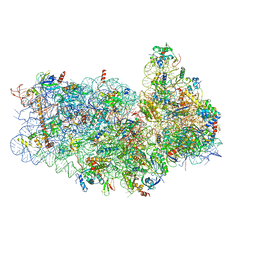 | | Structure of the yeast Kluyveromyces lactis small ribosomal subunit in complex with the cricket paralysis virus IRES. | | Descriptor: | 18S ribosomal RNA, Cricket paralysis virus IRES RNA, MAGNESIUM ION, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
8WTB
 
 | |
3DWR
 
 | |
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
