3GHE
 
 | |
3GFW
 
 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrolo-pyridin ligand | | Descriptor: | 1-(4-(4-(2-(isopropylsulfonyl)phenylamino)-1H-pyrrolo[2,3-b]pyridin-6-ylamino)-3-methoxyphenyl)piperidin-4-ol, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Choi, H, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3G77
 
 | | Bacterial cytosine deaminase V152A/F316C/D317G mutant | | Descriptor: | Cytosine deaminase, FE (III) ION | | Authors: | Stoddard, B, Zhao, L. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo.
Cancer Res., 69, 2009
|
|
3G88
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group P61 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Belardinelli, R, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3G8O
 
 | | Progesterone Receptor with bound Pyrrolidine 1 | | Descriptor: | N~2~-[4-cyano-3-(trifluoromethyl)phenyl]-N,N-dimethyl-N~2~-(2,2,2-trifluoroethyl)-L-alaninamide, Progesterone receptor, SULFATE ION | | Authors: | Thompson, S.K, Washburn, D.G, Madauss, K.P, Williams, S.P, Stewart, E.L. | | Deposit date: | 2009-02-12 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of orally-active, pyrrolidine-based progesterone receptor partial agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G8Z
 
 | |
3GIG
 
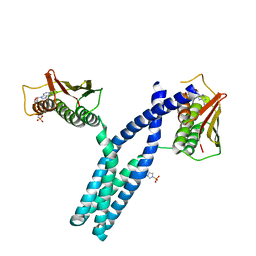 | | Crystal structure of phosphorylated DesKC in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GAS
 
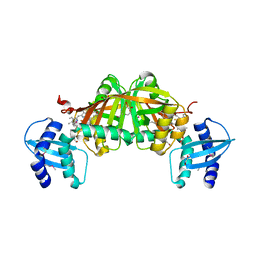 | | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, F, Hu, Y.L, Guo, Y, Guo, G, Zou, Q.M, Wang, D.C. | | Deposit date: | 2009-02-18 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme
To be Published
|
|
3GBH
 
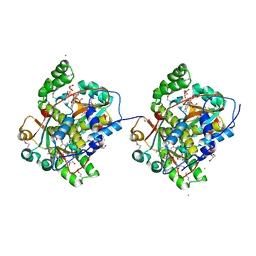 | |
3G8D
 
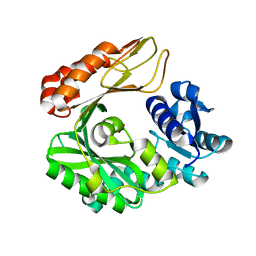 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
3GFI
 
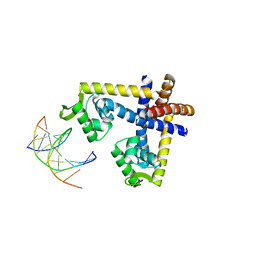 | | Crystal structure of ST1710 complexed with its promoter DNA | | Descriptor: | 146aa long hypothetical transcriptional regulator, 5'-D(*TP*AP*AP*CP*AP*AP*TP*AP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*GP*CP*TP*AP*TP*TP*GP*T)-3' | | Authors: | Kumarevel, T, Tanaka, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-02-26 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ST1710-DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators.
Nucleic Acids Res., 37, 2009
|
|
3G96
 
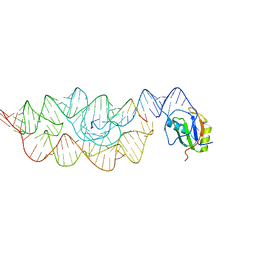 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3GA6
 
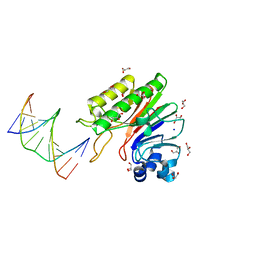 | | Mth0212 in complex with two DNA helices | | Descriptor: | 5'-D(*GP*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3GH2
 
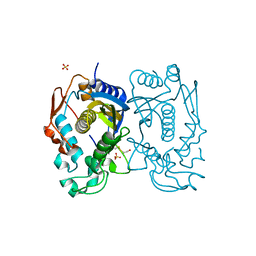 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3H0J
 
 | |
3H1O
 
 | | The Structure of Fluorescent Protein FP480 | | Descriptor: | Fluorescent protein FP480, GLYCEROL | | Authors: | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | Deposit date: | 2009-04-13 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GLB
 
 | | Crystal structure of the effector binding domain of a CATM variant (R156H) | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, GLYCEROL, HTH-type transcriptional regulator catM, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMG
 
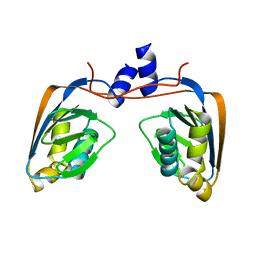 | | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv1825/MT1873 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis
To be Published
|
|
3GMS
 
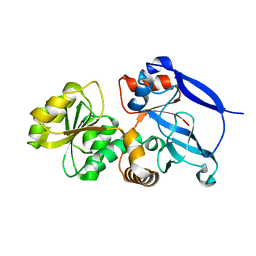 | |
3GO1
 
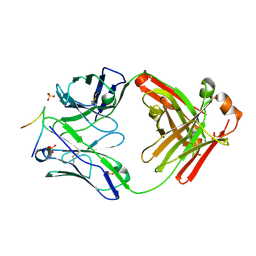 | |
3H36
 
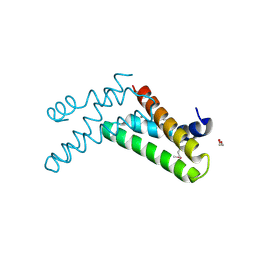 | | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3H3H
 
 | |
3GPX
 
 | | Sequence-matched MutM Interrogation Complex 4 (IC4) | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Spong, M.C, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
