3ITV
 
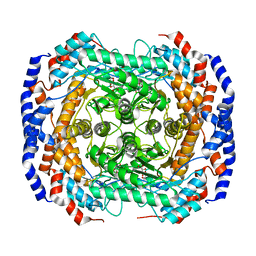 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUH
 
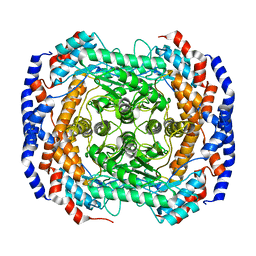 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COBALT (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3BY1
 
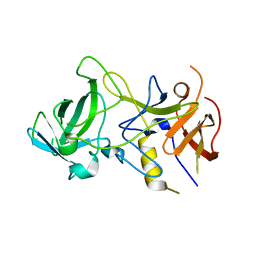 | | Unliganded Norvalk Virus P domain | | Descriptor: | 58 kd capsid protein | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-01-15 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
2PJJ
 
 | |
4OW0
 
 | |
3LD1
 
 | | Crystal Structure of IBV Nsp2a | | Descriptor: | Replicase polyprotein 1a | | Authors: | Xu, Y, Cong, L, Wei, L, Fu, J, Chen, C, Yang, A, Tang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2010-01-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | IBV nsp2 is an endosome-associated protein and viral pathogenicity factor
To be Published
|
|
3R3D
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant T77S complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3R36
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73Q complexed with 4-hydroxybenzoic acid | | Descriptor: | 4-hydroxybenzoyl-CoA thioesterase, P-HYDROXYBENZOIC ACID | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
2ZJF
 
 | |
4MKP
 
 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
3AHW
 
 | |
2QG6
 
 | |
3AB6
 
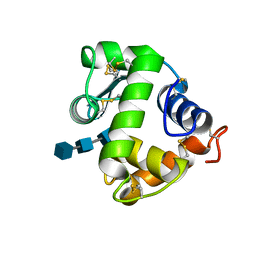 | | Crystal structure of NAG3 bound lysozyme from Meretrix lusoria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme | | Authors: | Yoneda, K, Kuwano, Y, Araki, T. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The tertiary structure of an i-type lysozyme isolated from the common orient clam (Meretrix lusoria)
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2QJH
 
 | |
3QVC
 
 | |
3D6G
 
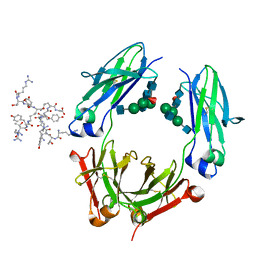 | | Fc fragment of IgG1 (Herceptin) with protein-A mimetic peptide dendrimer ligand. | | Descriptor: | 2-[[(2S)-2,6-bis[[(2S)-2,6-bis[[(2R)-2-[[(2R,3R)-2-[[(2R)-2-amino-5-carbamimidamido-pentanoyl]amino]-3-hydroxy-butanoyl]amino]-3-(4-hydroxyphenyl)propanoyl]amino]hexanoyl]amino]hexanoyl]amino]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Bujacz, A.D, Redzynia, I, Bujacz, G.D, Dinon, F, Pengo, P, Fassina, G. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of a Protein A mimetic peptide dendrimer bound to human IgG.
J.Phys.Chem.B, 113, 2009
|
|
4OVZ
 
 | |
3D26
 
 | | Norwalk P domain A-trisaccharide complex | | Descriptor: | 58 kd capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
2QJG
 
 | | M. jannaschii ADH synthase complexed with F1,6P | | Descriptor: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2QL6
 
 | | human nicotinamide riboside kinase (NRK1) | | Descriptor: | (1R)-1-[4-(AMINOCARBONYL)-1,3-THIAZOL-2-YL]-1,4-ANHYDRO-D-RIBITOL, ADENOSINE-5'-DIPHOSPHATE, nicotinamide riboside kinase 1 | | Authors: | Khan, J.A, Xiang, S, Tong, L. | | Deposit date: | 2007-07-12 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human nicotinamide riboside kinase
Structure, 15, 2007
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
5Z2B
 
 | |
1SEL
 
 | |
