1EPI
 
 | |
8CFZ
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H09 | | Descriptor: | 1-[2,4-bis(fluoranyl)phenyl]-2-(3,4-dihydro-1,2,4-triazol-2-yl)ethanone, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H09
To be published
|
|
1THP
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225P MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
8AV6
 
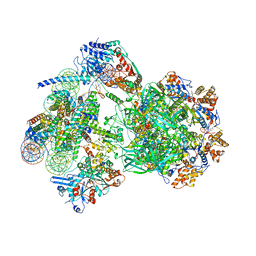 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8ATF
 
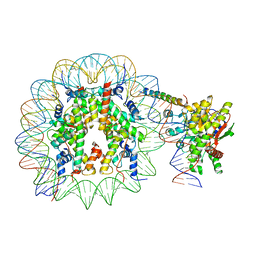 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1JT8
 
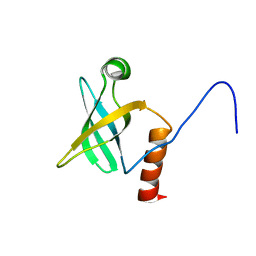 | | ARCHAEAL INITIATION FACTOR-1A, AIF-1A | | Descriptor: | PROBABLE TRANSLATION INITIATION FACTOR 1A | | Authors: | Hoffman, D.W, Li, W. | | Deposit date: | 2001-08-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of translation initiation factor aIF-1A from the archaeon Methanococcus jannaschii determined by NMR spectroscopy
Protein Sci., 10, 2001
|
|
7WIS
 
 | |
7BI5
 
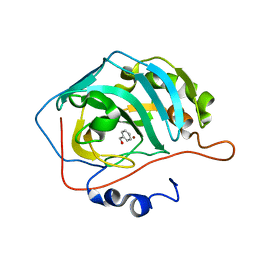 | | Human CA II in complex with benzyl alcohol | | Descriptor: | Carbonic anhydrase 2, ZINC ION, phenylmethanol | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Benzyl alcohol inhibits carbonic anhydrases by anchoring to the zinc coordinated water molecule.
Biochem.Biophys.Res.Commun., 548, 2021
|
|
7WIR
 
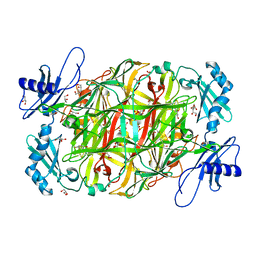 | |
5LHI
 
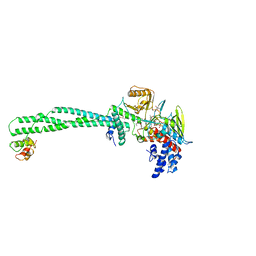 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
1F4Q
 
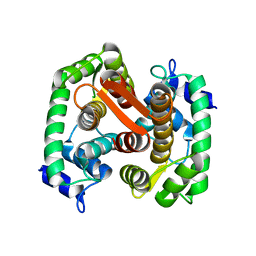 | | CRYSTAL STRUCTURE OF APO GRANCALCIN | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2000-06-08 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
3NAE
 
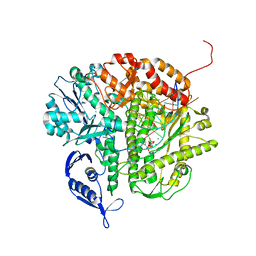 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite Guanidinohydantoin | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA Polymerase Allows dAMP and dGMP To Be Inserted opposite Guanidinohydantoin .
Biochemistry, 49, 2010
|
|
7TN2
 
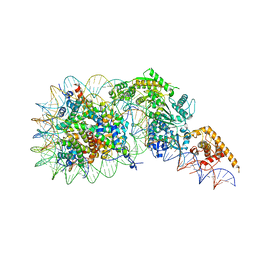 | | Composite model of a Chd1-nucleosome complex in the nucleotide-free state derived from 2.3A and 2.7A Cryo-EM maps | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1F4O
 
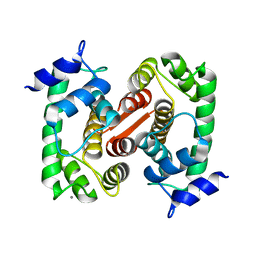 | | CRYSTAL STRUCTURE OF GRANCALCIN WITH BOUND CALCIUM | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2000-06-08 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
8D6J
 
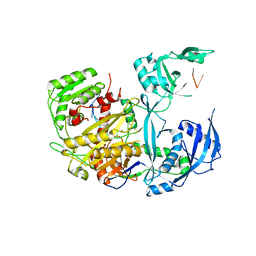 | |
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
8D71
 
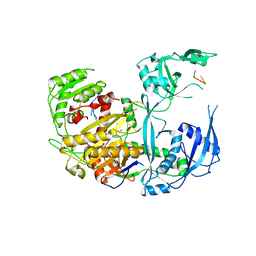 | | Human Ago2 bound to miR122(21nt) | | Descriptor: | MAGNESIUM ION, Protein argonaute-2, miR-122-21nt | | Authors: | Xiao, Y, MacRae, I. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tiny loop in the Argonaute PIWI domain tunes small RNA seed strength.
Embo Rep., 24, 2023
|
|
8D37
 
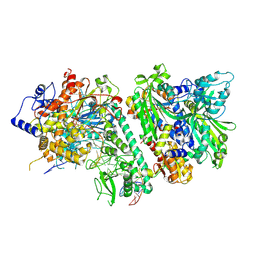 | |
6DIA
 
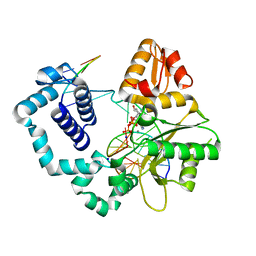 | | DNA polymerase beta substrate complex with templating cytosine and incoming Fapy-dGTP analog | | Descriptor: | 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
3RI1
 
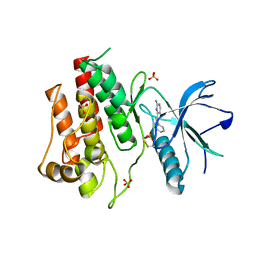 | | Crystal structure of the catalytic domain of FGFR2 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
8D3R
 
 | |
8D42
 
 | |
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
