3CE4
 
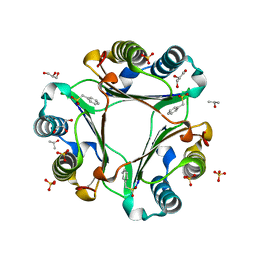 | |
1DQS
 
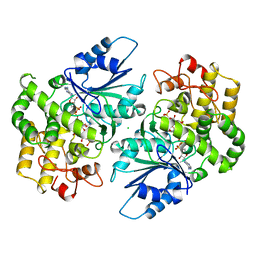 | | CRYSTAL STRUCTURE OF DEHYDROQUINATE SYNTHASE (DHQS) COMPLEXED WITH CARBAPHOSPHONATE, NAD+ AND ZN2+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-DEHYDROQUINATE SYNTHASE), ... | | Authors: | Carpenter, E.P, Hawkins, A.R, Frost, J.W, Brown, K.A. | | Deposit date: | 1998-04-09 | | Release date: | 1999-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis.
Nature, 394, 1998
|
|
1WV1
 
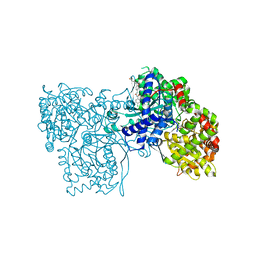 | | Crystallographic studies on acyl ureas, a new class of inhibitors of glycogenphosphorylase. Broad specificity of the allosteric site | | Descriptor: | 5-[3-({[(2,4-DICHLOROBENZOYL)AMINO]CARBONYL}AMINO)-2-METHYLPHENOXY]PENTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Klabunde, T, Wendt, K.U, Defossa, E. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
8IYB
 
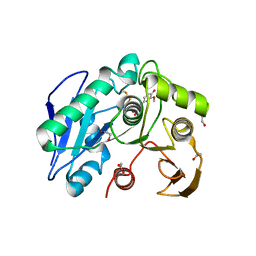 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IYC
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
1WUY
 
 | | Crystallographic studies on acyl ureas, a new class of inhibitors of glycogen phosphorylase. Broad specificity of the allosteric site | | Descriptor: | 4-[3-CHLORO-4-({[(2,4-DICHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]BUTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Klabunde, T, Wendt, K.U, Defossa, E. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
7NWA
 
 | |
7PIU
 
 | | Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
7PIV
 
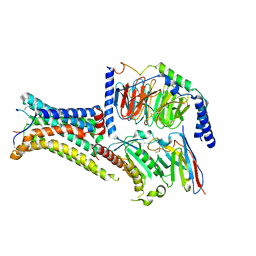 | | Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody VHH fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
7PUF
 
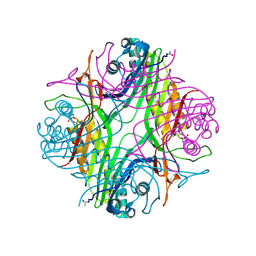 | | urate oxidase azaxanthine complex under 600 bar (60 MPa) of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
3L4O
 
 | |
7Q0N
 
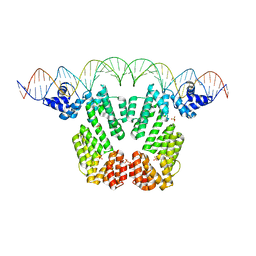 | | Arbitrium receptor from Katmira phage | | Descriptor: | Arbitrium receptor, DNA (45-MER), SULFATE ION | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of action of the arbitrium communication system in SPbeta phages.
Nat Commun, 13, 2022
|
|
5BQI
 
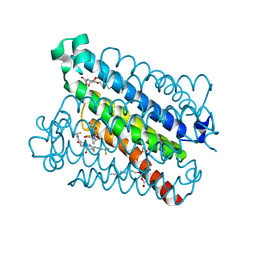 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}-N-{5-methyl-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-2-yl}pyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
4XUJ
 
 | |
5BQH
 
 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, GLUTATHIONE, N-[4-(4-chlorophenyl)-1H-imidazol-2-yl]-2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}benzamide, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5EBX
 
 | |
2R22
 
 | |
2W7Z
 
 | | Structure of the pentapeptide repeat protein EfsQnr, a DNA gyrase inhibitor. Free amines modified by cyclic pentylation with glutaraldehyde. | | Descriptor: | CHLORIDE ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Vetting, M.W, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2009-01-06 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of a Pentapeptide-Repeat Protein by Reductive Cyclic Pentylation of Free Amines with Glutaraldehyde.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ERA
 
 | | RECOMBINANT ERABUTOXIN A (S8T MUTANT) | | Descriptor: | ERABUTOXIN A, THIOCYANATE ION | | Authors: | Gaucher, J.F, Menez, R, Arnoux, B, Menez, A, Ducruix, A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution x-ray analysis of two mutants of a curaremimetic snake toxin
Eur.J.Biochem., 267, 2000
|
|
3RXG
 
 | | Crystal structure of Trypsin complexed with 4-aminocyclohexanol | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXT
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamin (F04 and F03, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXJ
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXC
 
 | | Crystal structure of Trypsin complexed with 2-aminopyridine | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXU
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and A06, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXQ
 
 | | Crystal structure of Trypsin complexed with benzamide (F01 and F05, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
