5NYF
 
 | |
2R51
 
 | | Crystal Structure of mouse Vps26B | | Descriptor: | Vacuolar protein sorting-associated protein 26B | | Authors: | Owen, D.J, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2007-09-02 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Vps26B and mapping of its interaction with the retromer protein complex.
Traffic, 9, 2008
|
|
4GCF
 
 | |
3UXJ
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ0 | | Descriptor: | 1,2-ETHANEDIOL, 7-DEAZA-7-AMINOMETHYL-GUANINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Zhang, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from
Vibrio cholerae complexed with NADP and PreQ0
To be Published, 2012
|
|
5A14
 
 | | Human CDK2 with type II inhibitor | | Descriptor: | 1-[4-(2-azanylpyrimidin-4-yl)oxyphenyl]-3-[4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]urea, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Alexander, L.T, Elkins, J.M, Kopec, J, Fedorov, O, Savitsky, P.A, Moebitz, H, Cowan-Jacob, S.W, Szklarz, M, Pike, A.C.W, Carpenter, E.P, Krojer, T, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2015-04-27 | | Release date: | 2015-07-22 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II Inhibitors Targeting Cdk2.
Acs Chem.Biol., 10, 2015
|
|
3V0G
 
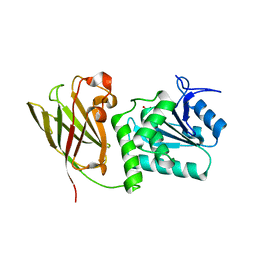 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form III | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5A1Q
 
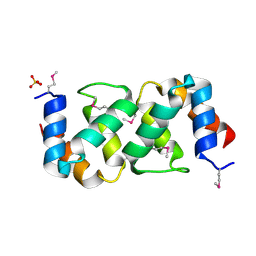 | |
2R8R
 
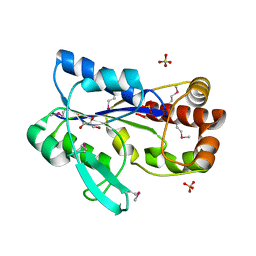 | | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3V13
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4GEW
 
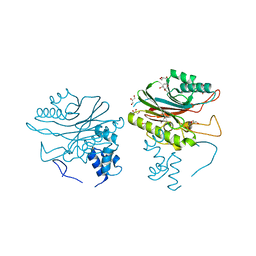 | |
4WE4
 
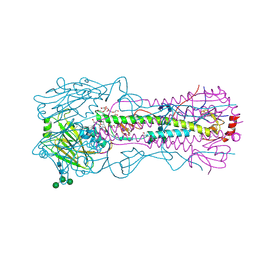 | | The crystal structure of hemagglutinin from 1968 H3N2 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
3V1H
 
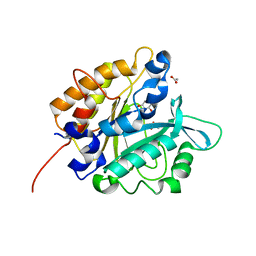 | | Structure of the H258Y mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
2R9A
 
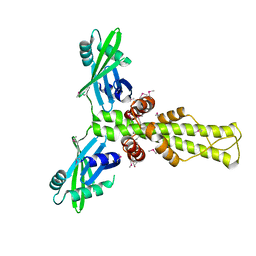 | | Crystal structure of human XLF | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Mol.Cell, 28
|
|
5O5O
 
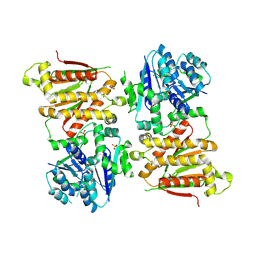 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
4FRX
 
 | |
3V4X
 
 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthase, mvaS, by Hymeglusin | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
5ACN
 
 | |
2RAQ
 
 | | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus. Northeast Structural Genomics Consortium target TT205 | | Descriptor: | CALCIUM ION, Conserved protein MTH889 | | Authors: | Forouhar, F, Su, M, Xu, X, Seetharaman, J, Mao, L, Xiao, R, Ma, L.-C, Conover, K, Baran, M.C, Acton, T.B, Montelione, G.T, Arrowsmith, C.H, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus.
To be Published
|
|
4FSP
 
 | |
5ADK
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((3-(Dimethylamino)methyl)phenoxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-[(dimethylamino)methyl]phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
3V5R
 
 | | Crystal structure of the unliganded form of Gal3p | | Descriptor: | Protein GAL3, SULFATE ION | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
4FT6
 
 | |
2RR4
 
 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
4GKV
 
 | | Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD | | Descriptor: | Alcohol dehydrogenase, propanol-preferring, GLYCEROL, ... | | Authors: | Sims, P.A, Thomas, L.M, Harper, A.R, Miner, W.A, Ajufo, H.O, Branscrum, K.M, Kao, L. | | Deposit date: | 2012-08-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
