9FXP
 
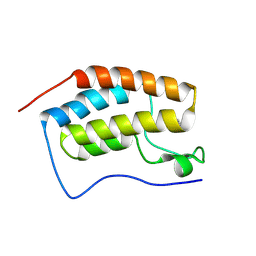 | | Crystal structure of BRD4 BD1 with DI00626383. | | Descriptor: | 4-methoxy-1,2-benzoxazol-3-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Reinert, D. | | Deposit date: | 2024-07-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
8YF7
 
 | |
8YF8
 
 | |
8YF9
 
 | |
6G6J
 
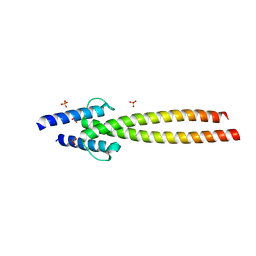 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6G6K
 
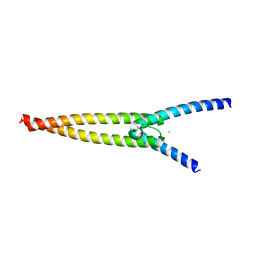 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | CHLORIDE ION, Myc proto-oncogene protein, Protein max | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
8GVN
 
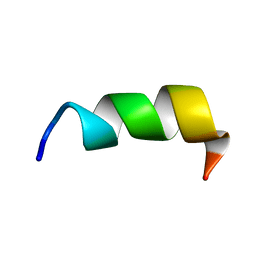 | | Novel salt-resistant antimicrobial peptide, RR14 | | Descriptor: | TRP-LEU-ARG-ARG-ILE-LYS-ALA-TRP-LEU-ARG-ARG-ILE-LYS-ALA | | Authors: | Lin, T.L, Tseng, T.S, Fan, P.J. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Deciphering Structure-Function Relationship Unveils Salt-Resistant Mode of Action of a Potent MRSA-Inhibiting Antimicrobial Peptide, RR14.
J.Bacteriol., 204, 2022
|
|
8G24
 
 | |
8G25
 
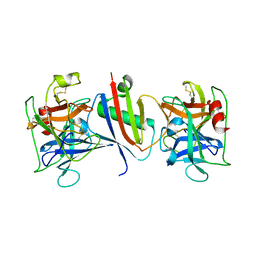 | |
2V1Y
 
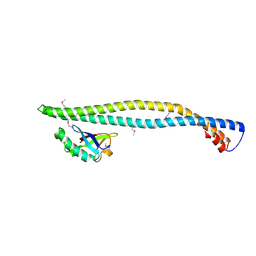 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
8G26
 
 | |
8GP7
 
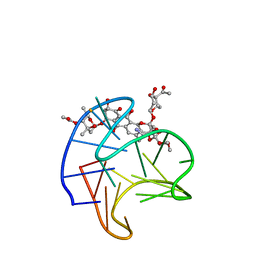 | | Structure of Trioxacarcin A covalently bound to RET G4-DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), Trioxacarcin A, bound form | | Authors: | Yin, S, Cao, C. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Trioxacarcin A Interactions with G-Quadruplex DNA Reveal Its Potential New Targets as an Anticancer Agent.
J.Med.Chem., 66, 2023
|
|
2W4U
 
 | | Isometrically contracting insect asynchronous flight muscle quick frozen after a length step | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, TROPOMYOSIN ALPHA-1 CHAIN, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-12-02 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Changes in Isometrically Contracting Insect Flight Muscle Trapped Following a Mechanical Perturbation.
Plos One, 7, 2012
|
|
8G9A
 
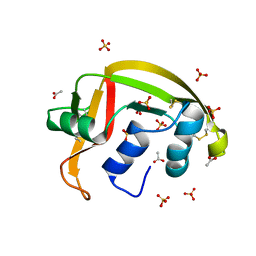 | | Crystal structure of a resurrected ancestor (AncRNase) of the pancreatic-type RNases 2 and 3 sub-families | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2023-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
2WJL
 
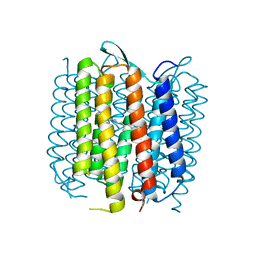 | | Bacteriorhodopsin mutant E194D | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Potschies, M, Wolf, S, Freier, E, Hofmann, E, Gerwert, K. | | Deposit date: | 2009-05-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
8HDJ
 
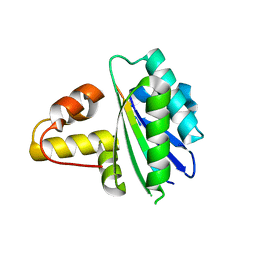 | |
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8HN2
 
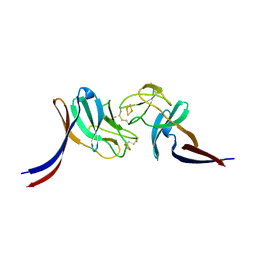 | | Selenomethionine-labelled soluble domain of Rieske iron-sulfur protein from chlorobaculum tepidum | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Kishimoto, H, Mutoh, R, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
6HQC
 
 | |
8FG6
 
 | |
8FTQ
 
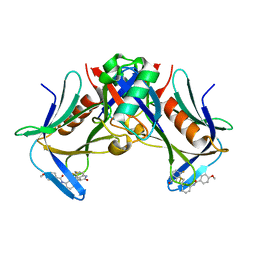 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
8HR4
 
 | |
2VWF
 
 | | Grb2 SH3C (2) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
8GGG
 
 | | RNase A-Adenosine 5'-Hexaphosphate (RNaseA.p6A) | | Descriptor: | GLYCEROL, Ribonuclease pancreatic, adenosine 5'-hexaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A.
Acs Cent.Sci., 10, 2024
|
|
8GE0
 
 | |
