5FYD
 
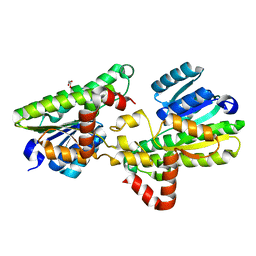 | | Structural and biochemical insights into 7beta-hydroxysteroid dehydrogenase stereoselectivity | | Descriptor: | GLYCEROL, OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | Authors: | Savino, S, Ferrandi, E, Forneris, F, Rovida, S, Riva, S, Monti, D, Mattevi, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Insights Into 7Beta-Hydroxysteroid Dehydrogenase Stereoselectivity.
Proteins, 84, 2016
|
|
7AJU
 
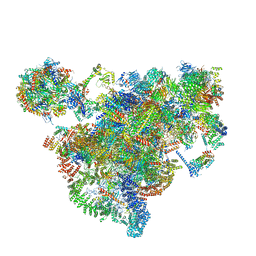 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7JVQ
 
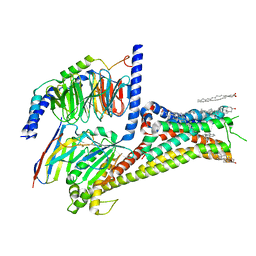 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
8R6Y
 
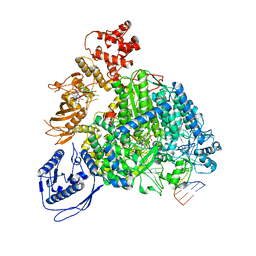 | | Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*(M7G)*AP*AP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
6E9Z
 
 | | DHF119 filament | | Descriptor: | DHF119 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
7JVP
 
 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7B53
 
 | | Crystal structure of MurE from E.coli | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5FCL
 
 | | Crystal structure of Cas1 from Pectobacterium atrosepticum | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Wilkinson, M.E, Nakatani, Y, Opel-Reading, H.K, Fineran, P.C, Krause, K.L. | | Deposit date: | 2015-12-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and in vivo activity of Cas1 from the type I-F CRISPR-Cas system.
Biochem.J., 473, 2016
|
|
7AV9
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
2I9P
 
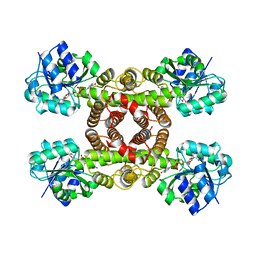 | | Crystal structure of human hydroxyisobutyrate dehydrogenase complexed with NAD+ | | Descriptor: | 3-hydroxyisobutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavanagh, K.L, Papagrigoriou, E, Salah, E, Lukacik, P, Smee, C, Burgess, N, von Delft, F, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-06 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of human hydroxyisobutyrate dehydrogenase complexed with NAD+
To be Published
|
|
6SBQ
 
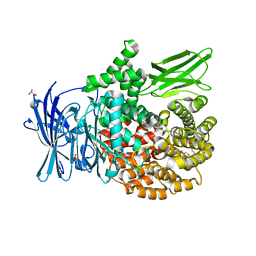 | | The crystal structure of PfA-M1 in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | DI(HYDROXYETHYL)ETHER, M1-family alanyl aminopeptidase, MALONATE ION, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
7AJT
 
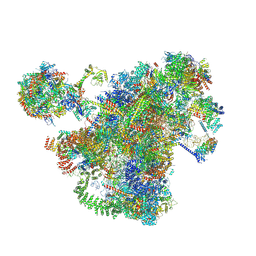 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
5DW7
 
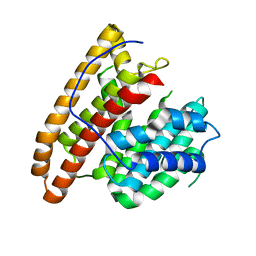 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
5FTG
 
 | | Human choline kinase a1 in complex with compound 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1- yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N- dimethyl-pyridin-4-amine (compound 10a) | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1-yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N-dimethyl-pyridin-4-amine, CHOLINE KINASE ALPHA | | Authors: | Schiaffino-Ortega, S, Baglioni, E, Mariotto, E, Bortolozzi, R, Serran-Aguilera, L, Rios-Marco, P, Carrasco-Jimenez, M.P, Gallo, M.A, Hurtado-Guerrero, R, Marco, C, Basso, G, Viola, G, Entrena, A, Lopez-Cara, L.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design, Synthesis, Crystallization and Biological Evaluation of New Symmetrical Biscationic Compounds as Selective Inhibitors of Human Choline Kinase Alpha1 (Chokalpha1)
Sci.Rep., 6, 2016
|
|
4TNN
 
 | | Crystal structure of Escherichia coli protein YodA in complex with Ni - artifact of purification. | | Descriptor: | Metal-binding lipocalin, NICKEL (II) ION, SULFATE ION | | Authors: | Gasiorowska, O.A, Cymborowski, M.T, Handing, K.B, Shabalin, I.G, Zasadzinska, E, Niedzialkowska, E, Porebski, P.J, Minor, W. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
1SD1
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1CY9
 
 | |
8F48
 
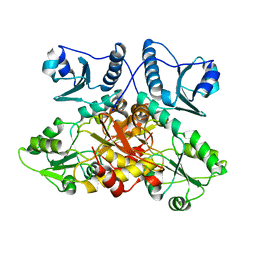 | |
7AHJ
 
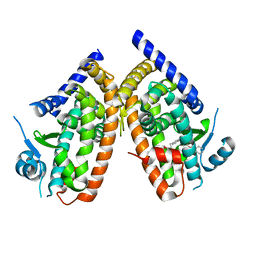 | | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar | | Descriptor: | 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Schoenmakers, E, Schwabe, B.T.W, Fairall, L, Chatterjee, K, Schwabe, J.W.R. | | Deposit date: | 2020-09-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar
To Be Published
|
|
4Y8Q
 
 | |
1SD2
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH 5'-METHYLTHIOTUBERCIDIN | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
4Y75
 
 | | Yeast 20S proteasome in complex with Ac-PAF-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAF-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
7BXT
 
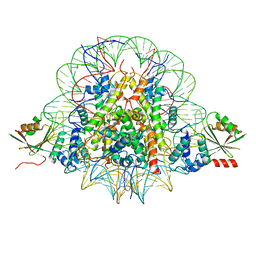 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
4Y95
 
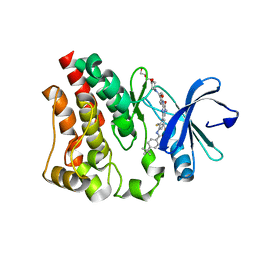 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Wang, Q, Rosen, C.E, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
1I7B
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND S-ADENOSYLMETHIONINE METHYL ESTER | | Descriptor: | 1,4-DIAMINOBUTANE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, S-ADENOSYLMETHIONINE DECARBOXYLASE BETA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
