3BH7
 
 | | Crystal structure of the RP2-Arl3 complex bound to GDP-AlF4 | | Descriptor: | ADP-ribosylation factor-like protein 3, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Veltel, S, Gasper, R, Wittinghofer, A. | | Deposit date: | 2007-11-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
2PMO
 
 | | Crystal structure of PfPK7 in complex with hymenialdisine | | Descriptor: | 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, Ser/Thr protein kinase | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
3B8Y
 
 | | Crystal Structure of Pectate Lyase PelI from Erwinia chrysanthemi in complex with tetragalacturonic acid | | Descriptor: | CALCIUM ION, Endo-pectate lyase, ZINC ION, ... | | Authors: | Creze, C, Castang, S, Derivery, E, Haser, R, Shevchik, V, Gouet, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia chrysanthemi in Complex with Its Substrate
J.Biol.Chem., 283, 2008
|
|
3BJQ
 
 | |
1OWD
 
 | | Substituted 2-Naphthamidine inhibitors of urokinase | | Descriptor: | 6-[AMINO(IMINO)METHYL]-N-[(4R)-4-ETHYL-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL]-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
3B9R
 
 | | SERCA Ca2+-ATPase E2 aluminium fluoride complex without thapsigargin | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Olesen, C, Picard, M, Winther, A.M.L, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of calcium transport by the calcium pump
Nature, 450, 2007
|
|
4B83
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-3-methoxy-benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
1OWK
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-1,2,3,4-TETRAHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
3C1A
 
 | |
3BSN
 
 | |
3BMZ
 
 | | Violacein biosynthetic enzyme VioE | | Descriptor: | Putative uncharacterized protein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-12-13 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Violacein Biosynthetic Enzyme VioE Shares a Fold with Lipoprotein Transporter Proteins
J.Biol.Chem., 283, 2008
|
|
2PKK
 
 | | Crystal structure of M tuberculosis Adenosine Kinase complexed with 2-fluro adenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
3BYQ
 
 | |
2HG1
 
 | |
4B3V
 
 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 20mM of Calcium Acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vaney, M.C, DuBois, R.M, Tortorici, M.A, Rey, F.A. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
1KQZ
 
 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
7XBZ
 
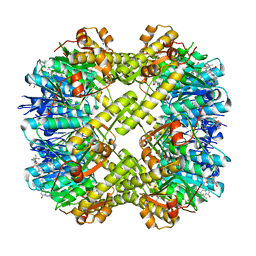 | | Crystal structure of Staphylococcus aureus ClpP in complex with R-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-[(1R)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
1F9D
 
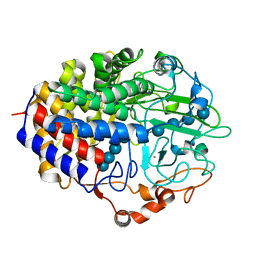 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellotetraose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
2Q55
 
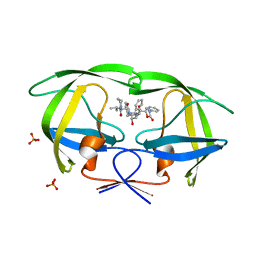 | | Crystal structure of KK44 bound to HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2S,4S)-1-BENZYL-2-HYDROXY-4-{[(2S)-3-METHYL-2-(2-OXOTETRAHYDROPYRIMIDIN-1(2H)-YL)BUTANOYL]AMINO}-5-PHENYLPENTYL]-2-OXO-3-PHENYL-1,3-OXAZOLIDINE-5-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2ZBS
 
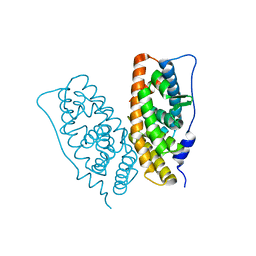 | |
2ZJN
 
 | |
1KPM
 
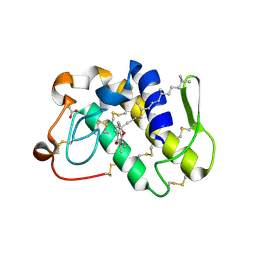 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
2ZAS
 
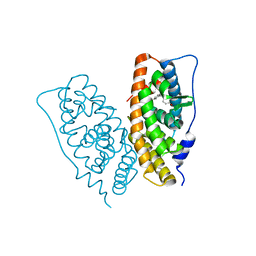 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2Q2N
 
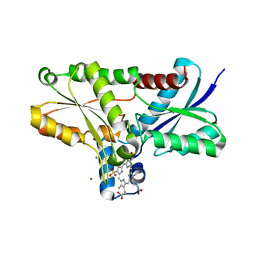 | | Crystal structure of Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
