7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
4HFN
 
 | |
1O8T
 
 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | Descriptor: | APOLIPOPROTEIN C-II | | Authors: | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | Deposit date: | 2002-11-29 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
1NRJ
 
 | |
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
5AA7
 
 | | Structural and functional characterization of a chitin-active 15.5 kDa lytic polysaccharide monooxygenase domain from a modular chitinase from Jonesia denitrificans | | Descriptor: | CHITINASE, COPPER (I) ION | | Authors: | Mekasha, S, Forsberg, Z, Dalhus, B, Choudhary, S, Schmidt-Dannert, C, Vaaje-Kolstad, G, Eijsink, V. | | Deposit date: | 2015-07-23 | | Release date: | 2015-12-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Small Chitin-Active Lytic Polysaccharide Monooxygenase Domain of a Multi-Modular Chitinase from Jonesia Denitrificans.
FEBS Lett., 590, 2016
|
|
1NSO
 
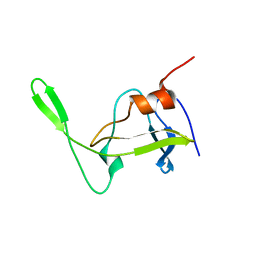 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
3V0O
 
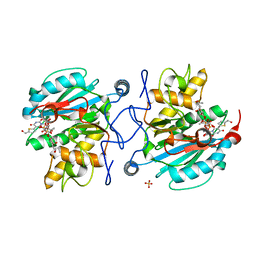 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with a novel UDP-GalNAc derived inhibitor (4GW) and H-antigen acceptor | | Descriptor: | 5-(5-formylthiophen-2-yl)uridine 5'-(trihydrogen diphosphate), Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Palcic, M.M, Jorgensen, R. | | Deposit date: | 2011-12-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Base-modified donor analogues reveal novel dynamic features of a glycosyltransferase.
J.Biol.Chem., 288, 2013
|
|
3V1K
 
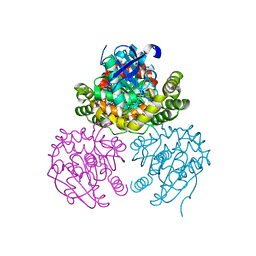 | |
7KSM
 
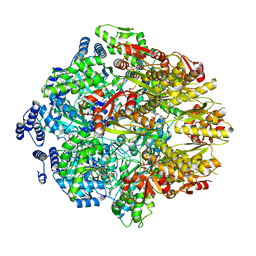 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
4P16
 
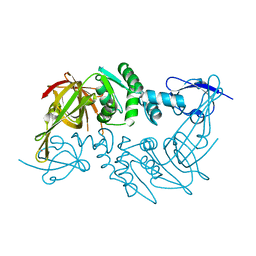 | | Crystal structure of the papain-like protease of Middle-East Respiratory Syndrome coronavirus | | Descriptor: | ORF1a, ZINC ION | | Authors: | Lei, J, Mesters, J.R, Ma, Q, Hilgenfeld, R. | | Deposit date: | 2014-02-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features.
Antiviral Res., 109C, 2014
|
|
1NU6
 
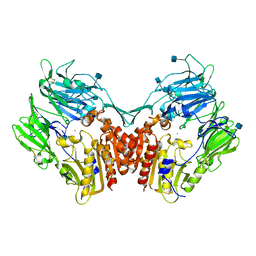 | | Crystal structure of human Dipeptidyl Peptidase IV (DPP-IV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, MERCURY (II) ION | | Authors: | Hennig, M, Stihle, M, Thoma, R, Ruf, A. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
4P1C
 
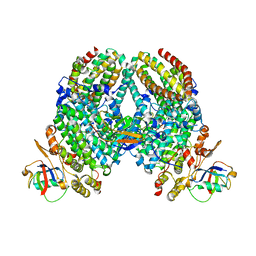 | | CRYSTAL STRUCTURE OF THE TOLUENE 4-MONOOXYGENASE HYDROXYLASE-FERREDOXIN C7S, C84A, C85A VARIANT ELECTRON-TRANSFER COMPLEX | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Acheson, J.F, Fox, B.G. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for biomolecular recognition in overlapping binding sites in a diiron enzyme system.
Nat Commun, 5, 2014
|
|
1O0S
 
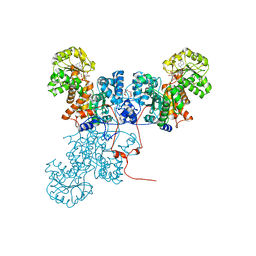 | | Crystal Structure of Ascaris suum Malic Enzyme Complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD-dependent malic enzyme, TARTRONATE | | Authors: | Rao, G.S, Coleman, D.E, Karsten, W.E, Cook, P.F, Harris, B.G. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
J.Biol.Chem., 278, 2003
|
|
2IFW
 
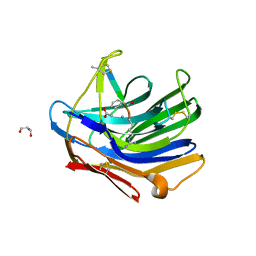 | | Crystal structure of scytalido-glutamic peptidase with a transition state analog inhibitor | | Descriptor: | ACETIC ACID, GLYCEROL, Heptapeptide, ... | | Authors: | Pillai, B, Cherney, M.M, Hiraga, K, Takada, K, Oda, K, James, M.N. | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis.
J.Mol.Biol., 365, 2007
|
|
7O2C
 
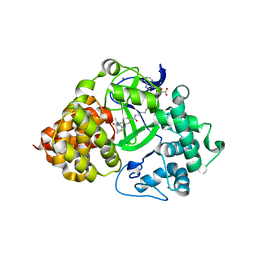 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH the benzodiazepine-based probe BAY-6035 | | Descriptor: | (2S)-1-[[(1R,5S)-3-azabicyclo[3.1.0]hexan-3-yl]carbonyl]-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Steuber, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
3V4Q
 
 | | Structure of R335W mutant of human Lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
3V4W
 
 | | Structure of E347K mutant of Lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
5AGD
 
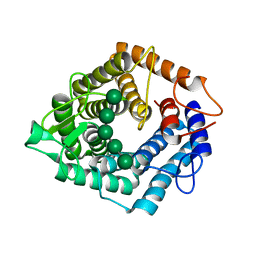 | | An inactive (D125N) variant of the catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with alpha-1,6-mannopentaose | | Descriptor: | ALPHA-1,6-MANNANASE, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Chandler, E, Temple, M.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7NY8
 
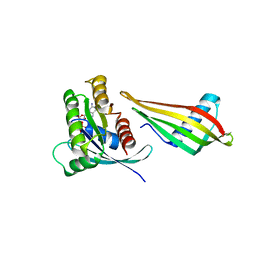 | | Affimer K69 - KRAS protein complex | | Descriptor: | Affimer K69, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turner, A.L, Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2021-03-21 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
7ZQ1
 
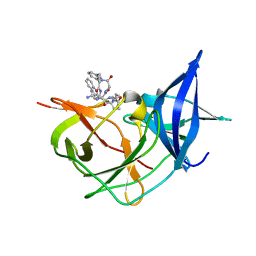 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2205 | | Descriptor: | 1-[(8R,15S,18S)-15-(4-azanylbutyl)-18-(1H-indol-3-ylmethyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-04-29 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
5AB0
 
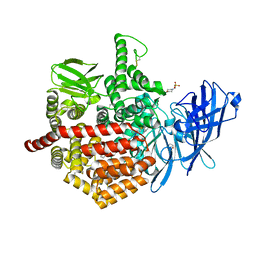 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
4P1X
 
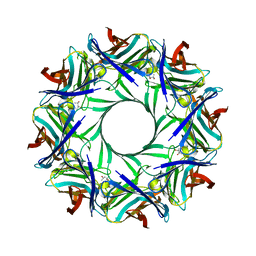 | | Crystal structure of staphylococcal LUK prepore | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component B, Gamma-hemolysin component C | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
5HYY
 
 | |
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
