8TJ4
 
 | | CRYSTAL STRUCTURE OF THE A/Bangkok/1/1979(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8ESW
 
 | | Structure of mitochondrial complex I from Drosophila melanogaster, Flexible-class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
5NBK
 
 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
8TJ9
 
 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
3L4O
 
 | |
8OVR
 
 | |
5MJ3
 
 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 1 | | Descriptor: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Hendrikx, P, Lasters, I, Savvides, S.N. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis Of Il-23 Antagonism By An Alphabody Protein Scaffold.
Nat Commun, 5, 2014
|
|
8TJA
 
 | | CRYSTAL STRUCTURE OF THE A/Ecuador/1374/2016(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
6ZQH
 
 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
6C1W
 
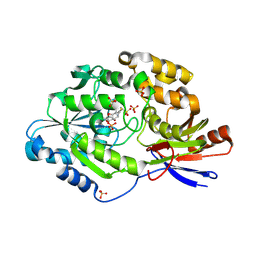 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemase, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-01-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Biochemistry, 57, 2018
|
|
8P1G
 
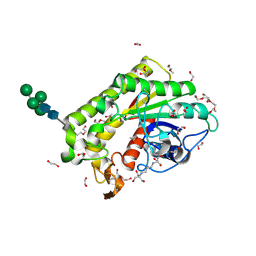 | | Crystal structure of inactive TtCE16 in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E, Karampa, P. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
8OTG
 
 | |
1BYJ
 
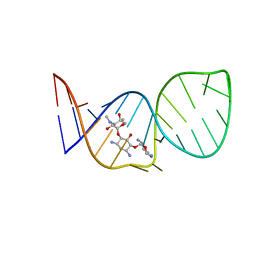 | | GENTAMICIN C1A A-SITE COMPLEX | | Descriptor: | 2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranose, 3,5-DIAMINO-CYCLOHEXANOL, 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranose, ... | | Authors: | Yoshizawa, S, Fourmy, D, Puglisi, J.D. | | Deposit date: | 1998-10-16 | | Release date: | 1999-10-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural origins of gentamicin antibiotic action.
EMBO J., 17, 1998
|
|
8OK2
 
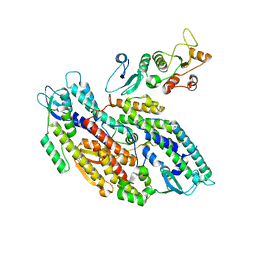 | | Bipartite interaction of TOPBP1 with the GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, DNA replication complex GINS protein PSF3, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2023-03-26 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | TopBP1 utilises a bipartite GINS binding mode to support genome replication.
Nat Commun, 15, 2024
|
|
8OTH
 
 | |
5N7T
 
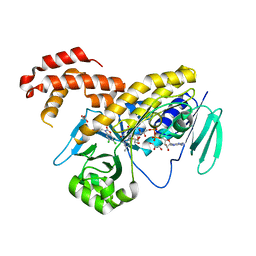 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5,6-dichloro-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The discovery of potent and selective kynurenine 3-monooxygenase inhibitors for the treatment of acute pancreatitis.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6FHV
 
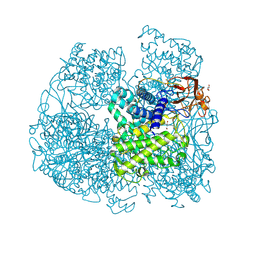 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F4Z
 
 | |
2O3V
 
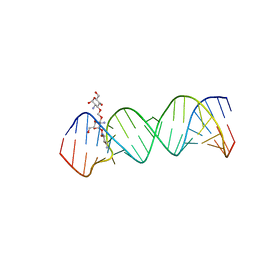 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O4K
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, CHLORIDE ION, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
3K0K
 
 | | Crystal Structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus. | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
7JL3
 
 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
8FWP
 
 | | Crystal Structure of CDC10 - CDC3 heterocomplex from Saccharomyces cerevisiae | | Descriptor: | Cell division control protein 10, Cell division control protein 3, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Silva, R.M, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
6BYX
 
 | | Complex structure of LOR107 mutant (R259N) with tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
