5NL7
 
 | | The crystal structure of the Actin Binding Domain (ABD) of alpha actinin from Entamoeba histolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative | | Authors: | Pinotsis, N, Djinovic-Carugo, K, Khan, M.B. | | Deposit date: | 2017-04-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4YX1
 
 | | SpaO(SPOA2) | | Descriptor: | CALCIUM ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
4ZRA
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS LPRG BINDING TO TRIACYLGLYCERIDE | | Descriptor: | Lipoprotein LprG, Tripalmitoylglycerol | | Authors: | Martinot, A.J, Farrow, M, Bai, L, Layre, E, Cheng, T.Y, Tsai, J.H.C, Iqbal, J, Annand, J, Sullivan, Z, Hussain, M, Sacchettini, J, Moody, D.B, Seeliger, J, Rubin, E.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis.
Plos Pathog., 12, 2016
|
|
7JIK
 
 | | Human recombinant Beta-2-Glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, ... | | Authors: | Klenotic, P.A, Yu, E.W.Y. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Beta-2-Glycoprotein I and it's role in APS
To Be Published
|
|
4OQX
 
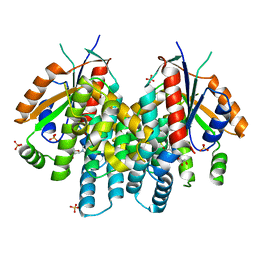 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with Me-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-methyl-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of HSV1-TK complexed with Me-ARA-EdU
To be Published
|
|
6SL3
 
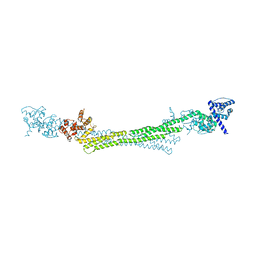 | |
6SL2
 
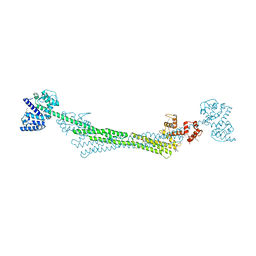 | | ALPHA-ACTININ FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative, ... | | Authors: | Pinotsis, N, Khan, M.B, Djinovic-Carugo, K. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SL7
 
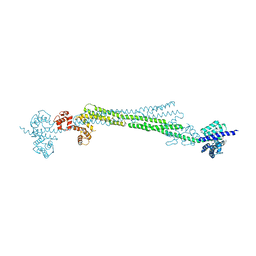 | |
1QQF
 
 | | N-TERMINALLY TRUNCATED C3D,G FRAGMENT OF THE COMPLEMENT SYSTEM | | Descriptor: | PROTEIN (COMPLEMENT C3DG) | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Berni, R. | | Deposit date: | 1999-06-04 | | Release date: | 2000-07-31 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
1QSJ
 
 | | N-TERMINALLY TRUNCATED C3DG FRAGMENT | | Descriptor: | COMPLEMENT C3 PRECURSOR | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Folli, C, Berni, R. | | Deposit date: | 1999-06-22 | | Release date: | 2000-07-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
1FH8
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH7
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FHD
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED IMIDAZOLE INHIBITOR | | Descriptor: | 5,6,7,8-TETRAHYDRO-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, BETA-1,4-XYLANASE, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH9
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED LACTAM OXIME INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-(2Z,3S,4S,5R)-2-hydroxyiminopiperidine-3,4,5-triol | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
3OQS
 
 | | Crystal structure of importin-alpha bound to a CLIC4 NLS peptide | | Descriptor: | Importin subunit alpha-2, peptide of Chloride intracellular channel protein 4 | | Authors: | Mynott, A.V, Brown, L.J, Harrop, S.J, Curmi, P.M.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of importin-alpha bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4
Febs J., 278, 2011
|
|
4CKD
 
 | |
5W1H
 
 | |
5W1I
 
 | |
5WLH
 
 | |
3BFC
 
 | | class A beta-lactamase SED-G238C complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Class A beta-lactamase Sed1 | | Authors: | Pernot, L, Petrella, S, Sougakoff, W. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acyl-intermediate structures of the class A beta-lactamase SED-G238C
To be Published
|
|
3BFF
 
 | | class A beta-lactamase SED-G238C complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Class A beta-lactamase Sed1, ... | | Authors: | Pernot, L, Petrella, S, Sougakoff, W. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | acyl-intermediate structures of the class A beta-lactamase SED-G238C
To be Published
|
|
3BFE
 
 | |
3BFG
 
 | | class A beta-lactamase SED-G238C complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class A beta-lactamase Sed1 | | Authors: | Pernot, L, Petrella, S, Sougakoff, W. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | acyl-intermediate structures of the class A beta-lactamase SED-G238C
To be Published
|
|
3BFD
 
 | |
4CC0
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
