4HXQ
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 14 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-carboxy-5-(methylamino)-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3H54
 
 | | Crystal Structure of human alpha-N-acetylgalactosaminidase,complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-04-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
4M6I
 
 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4LXB
 
 | | Crystal Structure Analysis of thrombin in complex with compound D58 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-difluoromethoxy-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-((S)-3-dimethylamino-pyrrolidin-1-yl)-3-oxo-propyl]-amide, Hirudin variant-1, ... | | Authors: | Stehlin-Gaon, C, Bocskei, Z. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | 5-Chlorothiophene-2-carboxylic acid [(S)-2-[2-methyl-3-(2-oxopyrrolidin-1-yl)benzenesulfonylamino]-3-(4-methylpiperazin-1-yl)-3-oxopropyl]amide (SAR107375), a selective and potent orally active dual thrombin and factor Xa inhibitor.
J.Med.Chem., 56, 2013
|
|
3MWV
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | Genome polyprotein | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
3GT5
 
 | | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa | | Descriptor: | CHLORIDE ION, N-acetylglucosamine 2-epimerase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa
To be Published
|
|
4MAH
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
3MWW
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | 1-[2-(4-carboxypiperidin-1-yl)-2-oxoethyl]-3-cyclohexyl-2-furan-3-yl-1H-indole-6-carboxylic acid, Genome polyprotein, SULFATE ION | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
4I7S
 
 | |
3MWB
 
 | | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A | | Descriptor: | MAGNESIUM ION, PHENYLALANINE, Prephenate dehydratase | | Authors: | Stein, A.J, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A
To be Published
|
|
3MX0
 
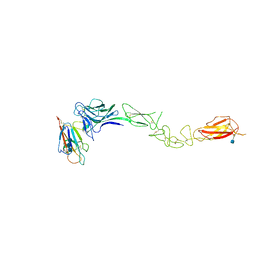 | | Crystal Structure of EphA2 ectodomain in complex with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 2, ... | | Authors: | Himanen, J.P, Yermekbayeva, L, Janes, P.W, Walker, J.R, Xu, K, Atapattu, L, Rajashankar, K.R, Mensinga, A, Lackmann, M, Nikolov, D.B, Dhe-Paganon, S. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4M2V
 
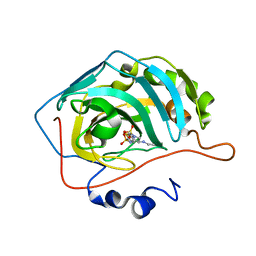 | | Genetically engineered Carbonic Anhydrase IX in complex with Brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
3MUX
 
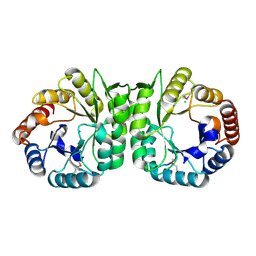 | | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A | | Descriptor: | CHLORIDE ION, SODIUM ION, putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Stein, A.J, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A
To be Published
|
|
3GZ5
 
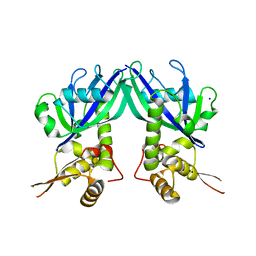 | | Crystal structure of Shewanella oneidensis NrtR | | Descriptor: | MutT/nudix family protein, SODIUM ION | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism
Structure, 17, 2009
|
|
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4I5G
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S, A86N, S88A | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4M8Z
 
 | |
3E2U
 
 | | Crystal structure of the zink-knuckle 2 domain of human CLIP-170 in complex with CAP-Gly domain of human dynactin-1 (p150-GLUED) | | Descriptor: | CAP-Gly domain-containing linker protein 1, Dynactin subunit 1, ZINC ION | | Authors: | Weisbrich, A, Honnappa, S, Capitani, G, Steinmetz, M.O. | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-function relationship of CAP-Gly domains
Nat.Struct.Mol.Biol., 14, 2007
|
|
4IDI
 
 | | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420 | | Descriptor: | GLYCEROL, Oryza sativa Rurm1-related | | Authors: | Wernimont, A.K, Tempel, W, Lew, J, Walker, J, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Bountra, C, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-12 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420
To be Published
|
|
4LTC
 
 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4MCC
 
 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
4IEL
 
 | | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione
To be Published
|
|
3E5H
 
 | | Crystal Structure of Rab28 GTPase in the Active (GppNHp-bound) Form | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lee, S.H, Baek, K, Li, Y, Dominguez, R. | | Deposit date: | 2008-08-13 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Large nucleotide-dependent conformational change in Rab28.
Febs Lett., 582, 2008
|
|
3EJS
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog: (5S)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5S,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
