3PM8
 
 | | CAD domain of PFF0520w, Calcium dependent protein kinase | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calcium-dependent protein kinase 2, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lew, J, Chamberlain, K, MacKenzie, F, Loppnau, P, Cossar, D, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CAD domain of PFF0520w, Calcium dependent protein kinase
TO BE PUBLISHED
|
|
4O6M
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
3PRD
 
 | |
4K0B
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4OGK
 
 | | X-ray structure of the uridine phosphorylase from Salmonella typhimurium in complex with thymidine at 2.40 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL, ... | | Authors: | Sotnichenko, S.E, Lashkov, A.A, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of the uridine phosphorylase from Salmonella typhimurium in complex with thymidine at 2.40 A resolution
To be Published
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3GW1
 
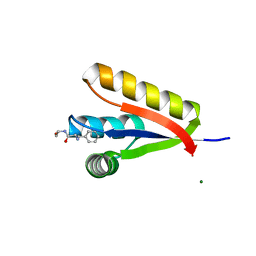 | | The structure of the Caulobacter crescentus CLPs protease adaptor protein in complex with FGG tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, FGG peptide, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4K3Q
 
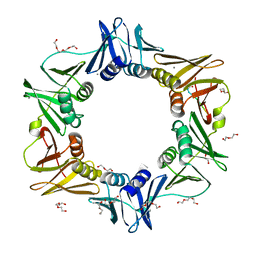 | | E. coli sliding clamp in complex with AcQLDAF | | Descriptor: | (ACE)QLDAF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4JZJ
 
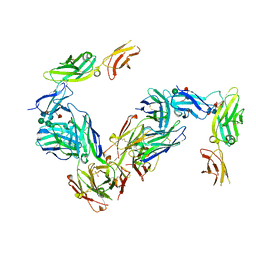 | | Crystal Structure of Receptor-Fab Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Broughton, S.E, Parker, M.W. | | Deposit date: | 2013-04-03 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody
Cell Rep, 8, 2014
|
|
3PU4
 
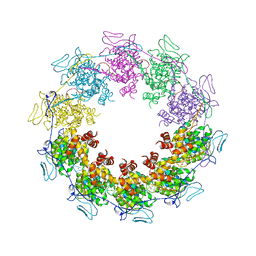 | |
3PV8
 
 | |
4K4T
 
 | | Poliovirus polymerase elongation complex (r4_form) | | Descriptor: | GLYCEROL, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*GP*AP*AP*A)-3'), RNA (5'-R(*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K60
 
 | | Crystal Structure of Human Chymase in Complex with Fragment 6-bromo-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromo-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
3OO6
 
 | |
4OE6
 
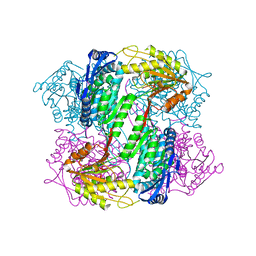 | | Crystal Structure of Yeast ALDH4A1 | | Descriptor: | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | | Authors: | Tanner, J.J. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Studies of Yeast Delta (1)-Pyrroline-5-carboxylate Dehydrogenase (ALDH4A1): Active Site Flexibility and Oligomeric State.
Biochemistry, 53, 2014
|
|
4OK3
 
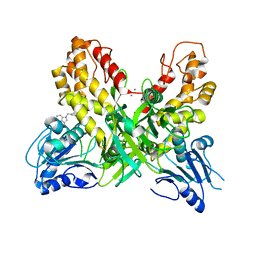 | |
3H05
 
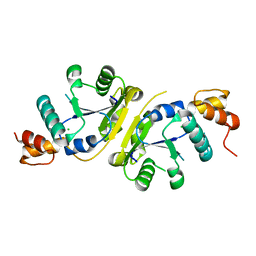 | | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, uncharacterized protein VPA0413 | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus
To be Published
|
|
4OFC
 
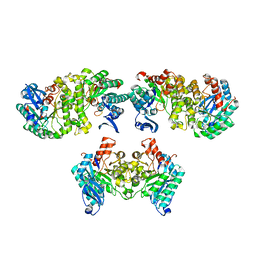 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
3H0V
 
 | | Human AdoMetDC with 5'-Deoxy-5'-(dimethylsulfonio) adenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-deoxy-5'-(dimethyl-lambda~4~-sulfanyl)adenosine, PYRUVIC ACID, ... | | Authors: | Bale, S, Brooks, W.H, Hanes, J.W, Mahesan, A.M, Guida, W.C, Ealick, S.E. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Role of the sulfonium center in determining the ligand specificity of human s-adenosylmethionine decarboxylase.
Biochemistry, 48, 2009
|
|
3H0B
 
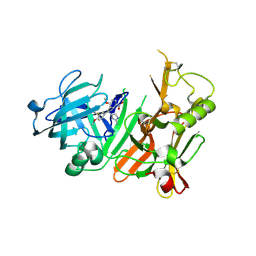 | | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class | | Descriptor: | 4-[(1S)-1-(3-fluoro-4-methoxyphenyl)-2-(2-methoxy-5-nitrophenyl)ethyl]-1H-imidazol-2-amine, Beta-secretase 1 | | Authors: | Allison, T.J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class: pH dependence on binding activity part 1.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4OGL
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution
Crystallogr. Rep., 2016
|
|
4K4Z
 
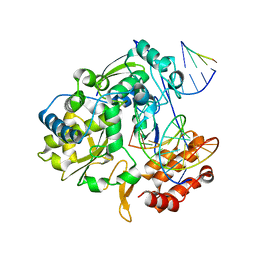 | | Coxsackievirus B3 polymerase elongation complex (r2_Mg_form) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4OHR
 
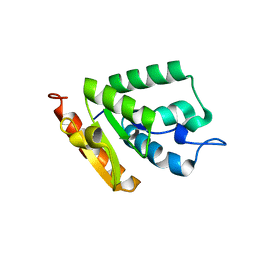 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
3H0X
 
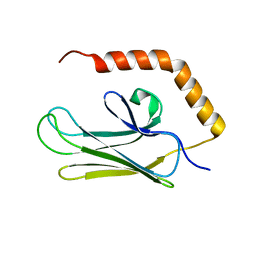 | | Crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae | | Descriptor: | 78 kDa glucose-regulated protein homolog | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae.
To be Published
|
|
4K6H
 
 | | Crystal structure of CALB mutant L278M from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
