2RKN
 
 | | X-ray structure of the self-defense and signaling protein DIR1 from Arabidopsis taliana | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, DIR1 protein, ZINC ION | | Authors: | Lascombe, M.B, Prange, T, Buhot, N, Marion, D, Bakan, B, Lamb, C. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein.
Protein Sci., 17, 2008
|
|
2RKU
 
 | | Structure of PLK1 in complex with BI2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Ding, Y.-H, Kothe, M, Kohls, D, Low, S. | | Deposit date: | 2007-10-17 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selectivity-determining residues in Plk1.
Chem.Biol.Drug Des., 70, 2007
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2RPN
 
 | | A crucial role for high intrinsic specificity in the function of yeast SH3 domains | | Descriptor: | Actin-binding protein, Actin-regulating kinase 1 | | Authors: | Stollar, E.J, Garcia, B, Chong, A, Forman-Kay, J, Davidson, A. | | Deposit date: | 2008-06-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p
J.Biol.Chem., 284, 2009
|
|
5BPC
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dATP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2RCX
 
 | | AmpC Beta-lactamase in complex with (1R)-1-(2-Thiophen-2-yl-acetylamino)-1-(3-(2-carboxyvinyl)-phenyl) methylboronic acid | | Descriptor: | (1R)-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-1-(3-(2-CARBOXYVINYL)-PHENYL) METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Morandi, F, Morandi, S, Prati, F, Shoichet, B.K. | | Deposit date: | 2007-09-20 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based optimization of cephalothin-analogue boronic acids as beta-lactamase inhibitors
Bioorg.Med.Chem., 16, 2008
|
|
2RDA
 
 | |
2RED
 
 | | Crystal structures of C2ALPHA-PI3 kinase PX-domain domain indicate conformational change associated with ligand binding. | | Descriptor: | GLYCEROL, Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide | | Authors: | Parkinson, G.N, Vines, D, Driscoll, P.C, Djordjevic, S. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding
Bmc Struct.Biol., 8, 2008
|
|
2D4A
 
 | | Structure of the malate dehydrogenase from Aeropyrum pernix | | Descriptor: | Malate dehydrogenase | | Authors: | Kawakami, R, Sakuraba, H, Tsuge, H, Ohshima, T. | | Deposit date: | 2005-10-12 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Refolding, characterization and crystal structure of (S)-malate dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
Biochim.Biophys.Acta, 1794, 2009
|
|
7AI5
 
 | | MutS in Scanning state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2RER
 
 | |
2RFO
 
 | | Crystral Structure of the nucleoporin Nic96 | | Descriptor: | Nucleoporin NIC96 | | Authors: | Schrader, N, Stelter, P, Flemming, D, Kunze, K, Hurt, E, Vetter, I.R. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the nic96 subcomplex organization in the nuclear pore channel.
Mol.Cell, 29, 2008
|
|
5BTS
 
 | |
7ANT
 
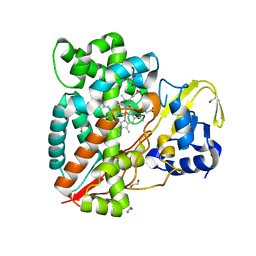 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
2RDJ
 
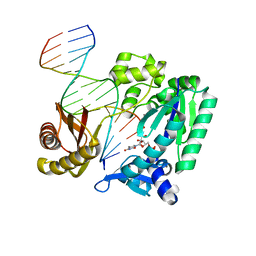 | |
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
2RH5
 
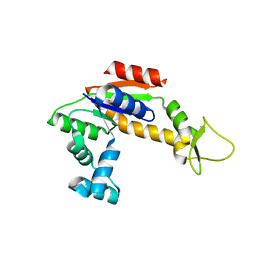 | | Structure of Apo Adenylate Kinase from Aquifex Aeolicus | | Descriptor: | Adenylate kinase | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
7AGH
 
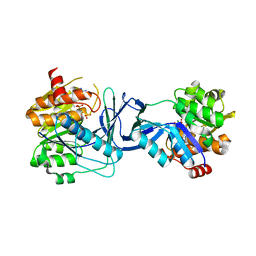 | |
7AG7
 
 | |
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AG4
 
 | |
2RFF
 
 | |
2RFK
 
 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7AGB
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Candidapepsin, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
