5D2F
 
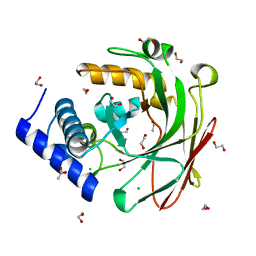 | | 4-oxalocrotonate decarboxylase from Pseudomonas putida G7 - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-oxalocrotonate decarboxylase NahK, ACETATE ION, ... | | Authors: | Guimaraes, S.L, Nagem, R.A.P. | | Deposit date: | 2015-08-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Crystal Structures of Apo and Liganded 4-Oxalocrotonate Decarboxylase Uncover a Structural Basis for the Metal-Assisted Decarboxylation of a Vinylogous beta-Keto Acid.
Biochemistry, 55, 2016
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
5D2J
 
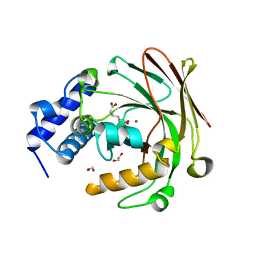 | |
2CC0
 
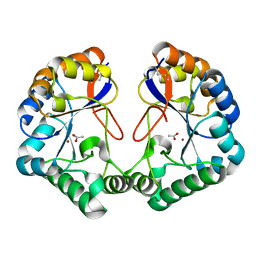 | | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate | | Descriptor: | ACETATE ION, ACETYL-XYLAN ESTERASE, ZINC ION | | Authors: | Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Vincent, F, Brzozowski, A.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Puchart, V, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases
J.Biol.Chem., 281, 2006
|
|
3FTT
 
 | | Crystal Structure of the galactoside O-acetyltransferase from Staphylococcus aureus | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Knapik, A.A, Shumilin, I.A, Cui, H, Xu, X, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
3FRK
 
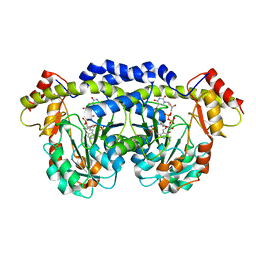 | | X-ray structure of QdtB from T. thermosaccharolyticum in complex with a PLP:TDP-3-aminoquinovose aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, QdtB | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose.
Biochemistry, 48, 2009
|
|
3FSY
 
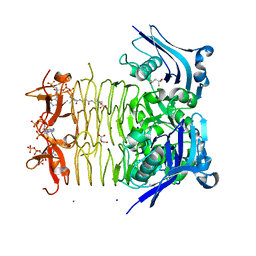 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c;DapD) in complex with succinyl-CoA from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
4ADL
 
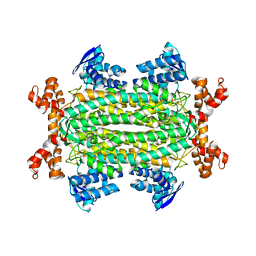 | | Crystal structures of Rv1098c in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
1JDI
 
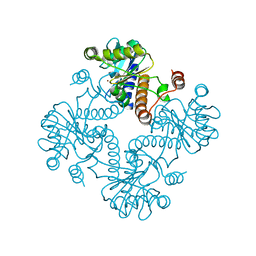 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
3AUU
 
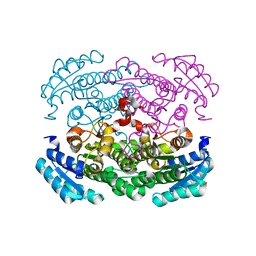 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | Descriptor: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
1T6R
 
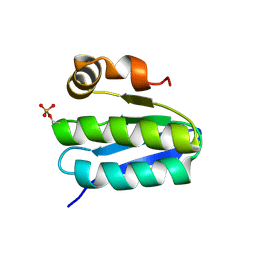 | | Solution structure of TM1442, a putative anti sigma factor antagonist in phosphorylated state | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjani, T, Placzek, W, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-05-07 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
3P2S
 
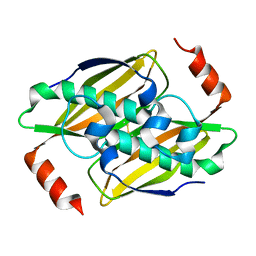 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in an open conformation | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
6UZB
 
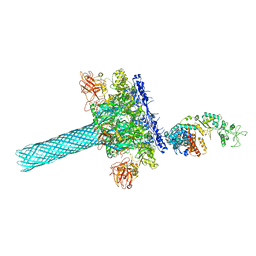 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
4A3X
 
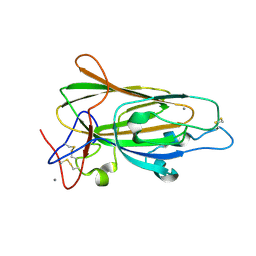 | | Structure of the N-terminal domain of the Epa1 adhesin (Epa1-Np) from the pathogenic yeast Candida glabrata, in complex with calcium and lactose | | Descriptor: | CALCIUM ION, EPA1P, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ielasi, F.S, Decanniere, K, Willaert, R.G. | | Deposit date: | 2011-10-05 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Epithelial Adhesin 1 (Epa1P) from the Human-Pathogenic Yeast Candida Glabrata : Structural and Functional Study of the Carbohydrate-Binding Domain
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ATQ
 
 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
5FFG
 
 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
3B3D
 
 | | B.subtilis YtbE | | Descriptor: | CALCIUM ION, Putative morphine dehydrogenase | | Authors: | Zhou, Y.F, Li, L.F, Liang, Y.H, Su, X.-D. | | Deposit date: | 2007-10-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3AY7
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | Descriptor: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
5D2H
 
 | |
4APA
 
 | |
3G8Q
 
 | |
4A15
 
 | | Crystal structure of an XPD DNA complex | | Descriptor: | 5'-D(*DTP*AP*CP*GP)-3', ATP-DEPENDENT DNA HELICASE TA0057, CALCIUM ION, ... | | Authors: | Kuper, J, Wolski, S.C, Michels, G, Kisker, C. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and Structural Studies of the Nucleotide Excision Repair Helicase Xpd Suggest a Polarity for DNA Translocation.
Embo J., 31, 2011
|
|
3FSX
 
 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c; DapD) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
8T4M
 
 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
4ADM
 
 | | Crystal structure of Rv1098c in complex with meso-tartrate | | Descriptor: | FUMARATE HYDRATASE CLASS II, GLYCEROL, S,R MESO-TARTARIC ACID | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-27 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
