4JTF
 
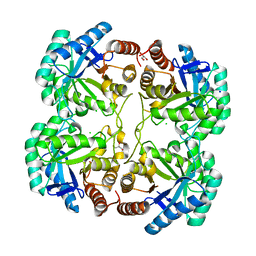 | | Crystal structure of F114R mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JOQ
 
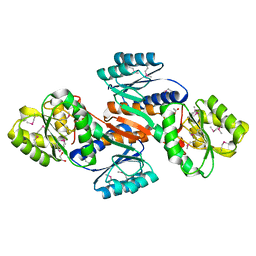 | | Putative ribose ABC transporter, periplasmic solute-binding protein from Rhodobacter sphaeroides | | Descriptor: | 1,2-ETHANEDIOL, ABC ribose transporter, periplasmic solute-binding protein, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putative ribose ABC transporter, periplasmic solute-binding protein from Rhodobacter sphaeroides.
To be Published
|
|
3NJI
 
 | | S114A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
4JRT
 
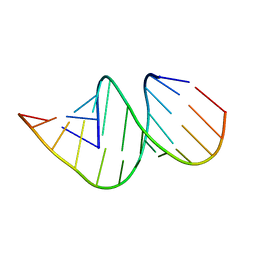 | | Crystal structure of an A-form RNA duplex containing three GU base pairs | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*CP*AP*CP*UP*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*UP*GP*GP*UP*GP*CP*GP*GP*G)-3') | | Authors: | Kondo, J, Dock-Bregeon, A.C, Willkomm, D.K, Hartmann, R.K, Westhof, E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an A-form RNA duplex obtained by degradation of 6S RNA in a crystallization droplet
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3G83
 
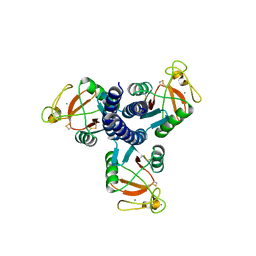 | |
4NLP
 
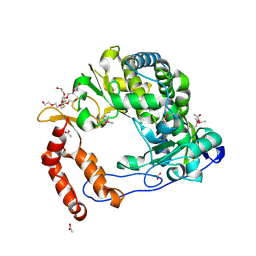 | | Poliovirus Polymerase - C290V Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4JVY
 
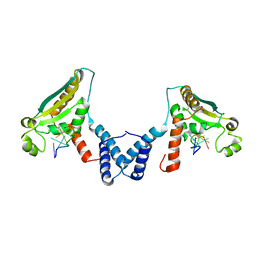 | | Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA | | Descriptor: | Female germline-specific tumor suppressor gld-1, RNA (5'-R(P*CP*UP*AP*AP*CP*AP*A)-3') | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
4NLX
 
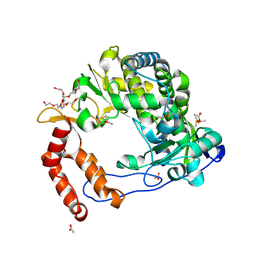 | | Poliovirus Polymerase - G289A/C290V Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NHV
 
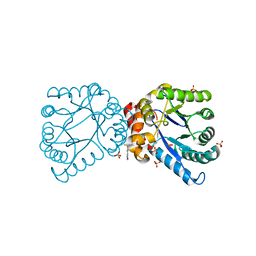 | |
4NMD
 
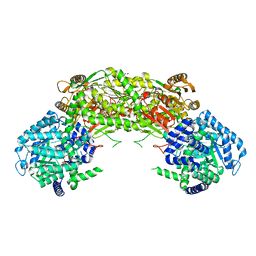 | |
3G8S
 
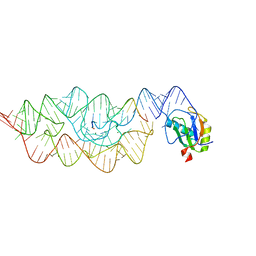 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4NO2
 
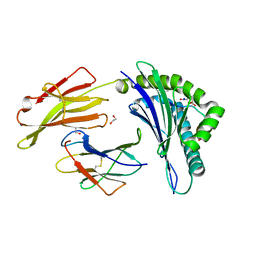 | | Crystal structure of RQA_V phosphopeptide bound to HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
4NIL
 
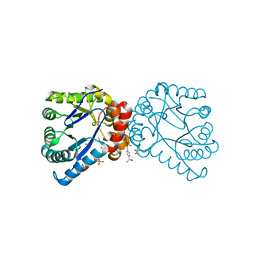 | |
3O9H
 
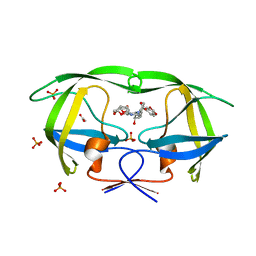 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3OAP
 
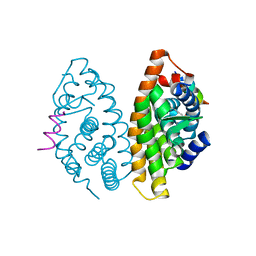 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9-cis retinoic acid and the coactivator peptide GRIP-1 | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, Energetics and Dynamics of Binding Coactivator Peptide to Human Retinoid X Receptor Alpha Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry, 50, 2011
|
|
4JUQ
 
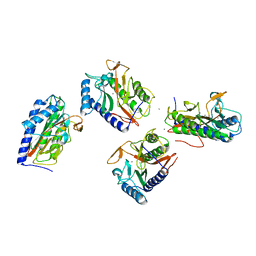 | | Pseudomonas aeruginosa MetAP T2N mutant, in Mn form | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2013-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methionine excision captured by the structures of a methionine aminopeptidase
To be published
|
|
3FQF
 
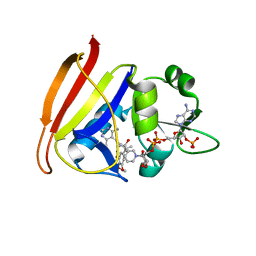 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(3,4,5-trimethoxyphenyl)pent-1-ynyl]-6-methylpyrimidine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
3GDS
 
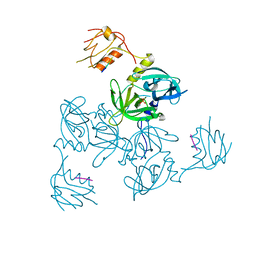 | |
3FQX
 
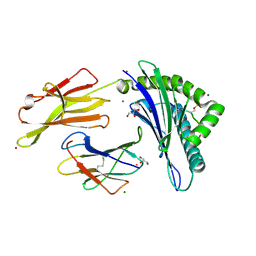 | |
3FSI
 
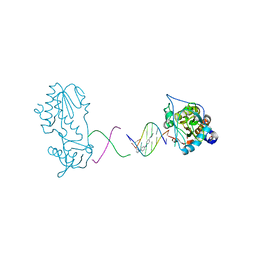 | | Crystal structure of a trypanocidal 4,4'-Bis(imidazolinylamino)diphenylamine bound to DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', ACETATE ION, ... | | Authors: | Glass, L.S, Georgiadis, M.M, Goodwin, K.D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a trypanocidal 4,4'-bis(imidazolinylamino)diphenylamine bound to DNA.
Biochemistry, 48, 2009
|
|
4NMC
 
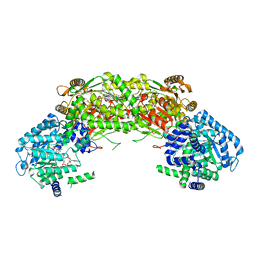 | |
3FSU
 
 | | Crystal Structure of Escherichia coli Methylenetetrahydrofolate Reductase Double Mutant Phe223LeuGlu28Gln complexed with methyltetrahydrofolate | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli.
Biochemistry, 48, 2009
|
|
4JCH
 
 | | OSH4 bound to an electrophilic oxysterol | | Descriptor: | (3beta,9beta,22R,25R)-3-hydroxyfurost-5-en-27-yl propanoate, Protein KES1 | | Authors: | Koag, M.C, Kou, Y.I, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-02-21 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of OSH4 bound to an electrophilic oxysterol
To be Published
|
|
3NT3
 
 | | CRYSTAL STRUCTURE OF LSSmKate2 red fluorescent proteins with large Stokes shift | | Descriptor: | GLYCEROL, LSSmKate2 red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V. | | Deposit date: | 2010-07-02 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering ESPT Pathways Based on Structural Analysis of LSSmKate Red Fluorescent Proteins with Large Stokes Shift.
J.Am.Chem.Soc., 132, 2010
|
|
4JFQ
 
 | | A2 HLA complex with L8A heteroclitic variant of Melanoma peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K, Baker, B.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T-cell receptor specificity maintained by altered thermodynamics.
J.Biol.Chem., 288, 2013
|
|
