7SLK
 
 | | [U:Ag+:U--pH 9] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
4W4O
 
 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
4WEI
 
 | | Crystal structure of the F4 fimbrial adhesin FaeG in complex with lactose | | Descriptor: | K88 fimbrial protein AD, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZVF
 
 | | Crystal structure of GGDEF domain of the E. coli DosC - form II (GTP-alpha-S-bound) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Diguanylate cyclase DosC, ... | | Authors: | Tarnawski, M, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural analysis of an oxygen-regulated diguanylate cyclase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6PCR
 
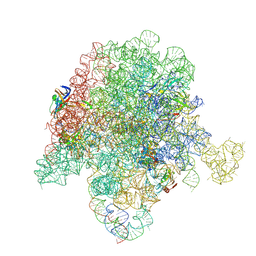 | | E. coli 50S ribosome bound to compound 40o | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl (2-bromopyridin-4-yl)carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5O30
 
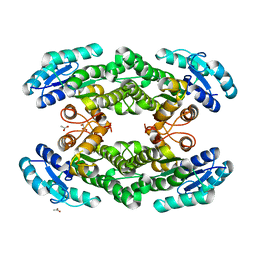 | |
5NTC
 
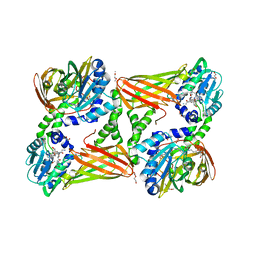 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0678 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2017-04-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0678
To Be Published
|
|
5A98
 
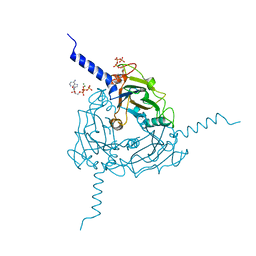 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5O4W
 
 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NVE
 
 | | Crystal structure of TNKS2 in complex with 2-(4-ethoxyphenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-(4-ethoxyphenyl)-3~{H}-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-05-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 2-Phenylquinazolinones as dual-activity tankyrase-kinase inhibitors.
Sci Rep, 8, 2018
|
|
5WFJ
 
 | | THE JAK3 KINASE DOMAIN IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 4-({[3-(propanoylamino)phenyl]methyl}amino)pyrrolo[1,2-b]pyridazine-3-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Sack, J. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of highly potent, selective, covalent inhibitors of JAK3.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5NWB
 
 | | Crystal structure of TNKS2 in complex with 2-{4-[(2-hydroxyethyl)(methyl)amino]phenyl}-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-[4-[2-hydroxyethyl(methyl)amino]phenyl]-3~{H}-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-05-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2-Phenylquinazolinones as dual-activity tankyrase-kinase inhibitors.
Sci Rep, 8, 2018
|
|
6P4H
 
 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
5NWK
 
 | | 14-3-3c in complex with CPP and fusicoccin | | Descriptor: | 14-3-3 c-1 protein, FUSICOCCIN, PHOSPHOSERINE, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
5O7A
 
 | | Quinolin-6-yloxyacetamides are microtubule destabilizing agents that bind to the colchicine site of tubulin | | Descriptor: | (2~{R})-2-(3-ethynylquinolin-6-yl)oxy-2-methoxy-~{N}-[(1~{E})-1-methoxyimino-2-methyl-propan-2-yl]ethanamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Olieric, N, Steinmetz, M.O. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Quinolin-6-Yloxyacetamides Are Microtubule Destabilizing Agents That Bind to the Colchicine Site of Tubulin.
Int J Mol Sci, 18, 2017
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
4ZWM
 
 | | 2.3 A resolution crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 | | Descriptor: | Ornithine aminotransferase, mitochondrial, putative, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Van Le, H, Silverman, R.B, McLeod, R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3 A resolution crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49
To Be Published
|
|
5NS2
 
 | | Cys-Gly dipeptidase GliJ in complex with Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
2BJC
 
 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | Authors: | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
