4LFK
 
 | |
4LH0
 
 | |
4IKY
 
 | | Crystal structure of peptide transporter POT (E310Q mutant) in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3N77
 
 | | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2 | | Descriptor: | Nucleoside triphosphatase nudI | | Authors: | Frydrysiak, J, Cooper, D.R, Derewenda, U, Anderson, W.F, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2
To be Published
|
|
3N7X
 
 | |
4LGB
 
 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
6ODM
 
 | | Herpes simplex virus type 1 (HSV-1) portal vertex-adjacent capsid/CATC, asymmetric unit | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
4INE
 
 | |
3GDU
 
 | |
3MV5
 
 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, GSK3-beta peptide, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-02 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
3MVJ
 
 | | Human cyclic AMP-dependent protein kinase PKA inhibitor complex | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pandit, J, Vajdos, F. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3GEW
 
 | | FaeE-FaeG chaperone-major pilin complex of F4 ad fimbriae | | Descriptor: | Chaperone protein faeE, GLYCEROL, K88 fimbrial protein AD, ... | | Authors: | Van Molle, I, Moonens, K, Garcia-Pino, A, Buts, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
J.Mol.Biol., 394, 2009
|
|
3N4G
 
 | | Crystal structure of native Cg10062 | | Descriptor: | Putative tautomerase | | Authors: | Guo, Y, Robertson, B.A, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-05-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of the Native and Inactivated Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase from Corynebacterium glutamicum: Implications for the Evolution of cis-3-Chloroacrylic Acid Dehalogenase Activity in the Tautomerase Superfamily
To be Published
|
|
4IR7
 
 | | Crystal Structure of Mtb FadD10 in Complex with Dodecanoyl-AMP | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, Long chain fatty acid CoA ligase FadD10, MAGNESIUM ION | | Authors: | Liu, Z, Wang, F, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis FadD10 protein reveal a new type of adenylate-forming enzyme.
J.Biol.Chem., 288, 2013
|
|
3GJB
 
 | | CytC3 with Fe(II) and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CytC3, ... | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
4LFC
 
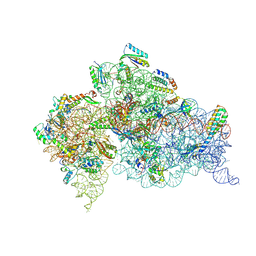 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: |
to be published, 2013
|
|
4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | Descriptor: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4ISA
 
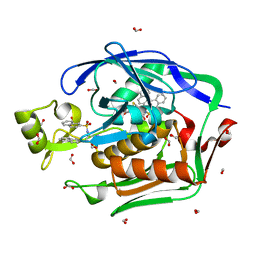 | | Crystal Structure of the Escherichia coli LpxC/BB-78485 complex | | Descriptor: | (2R)-N-hydroxy-3-naphthalen-2-yl-2-[(naphthalen-2-ylsulfonyl)amino]propanamide, (4S,5S)-1,2-DITHIANE-4,5-DIOL, FORMIC ACID, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
3MY7
 
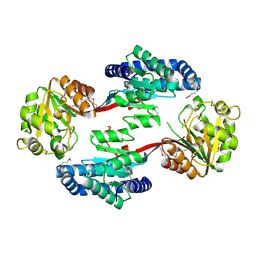 | | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A | | Descriptor: | Alcohol dehydrogenase/acetaldehyde dehydrogenase, CHLORIDE ION | | Authors: | Stein, A.J, Weger, A, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-10 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A
To be Published
|
|
4IT6
 
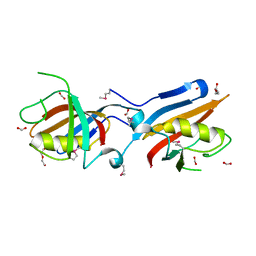 | |
4ITB
 
 | | Structure of bacterial enzyme in complex with cofactor and substrate | | Descriptor: | 1,2-ETHANEDIOL, 4-oxobutanoic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rhee, S, Park, J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-24 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
4LG5
 
 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
3MZY
 
 | |
4IU6
 
 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
