6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
7P84
 
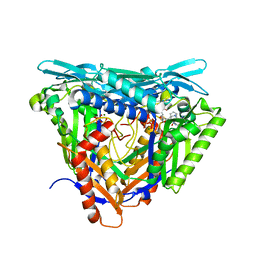 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P8M
 
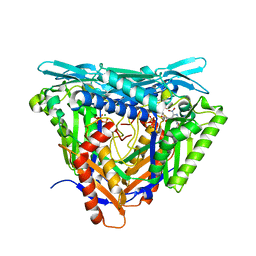 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with DMNB-SAM (4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | 4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine, MAGNESIUM ION, S-adenosylmethionine synthase, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
8TWL
 
 | |
4YB6
 
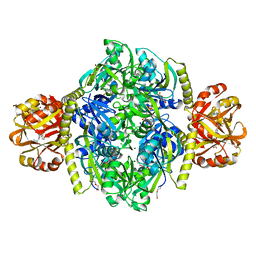 | | Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni in complex with the inhibitors AMP and histidine | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP phosphoribosyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mittelstaedt, G, Moggre, G.-J, Parker, E.J. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Campylobacter jejuni adenosine triphosphate phosphoribosyltransferase is an active hexamer that is allosterically controlled by the twisting of a regulatory tail.
Protein Sci., 25, 2016
|
|
7PBJ
 
 | |
8TWK
 
 | |
7QAN
 
 | | Cytochrome P450 Enzyme AbyV | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Parnell, A.E, Back, C.R, Race, P.R. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Role of Cytochrome P450 AbyV in the Final Stages of Abyssomicin C Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NCU
 
 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5K15
 
 | | Crystal structure of laccase from Thermus thermophilus HB27 (Cu2PO, 8 min) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Laccase | | Authors: | Diaz-Vilchis, A, Ruiz-Arellano, R.R, Rosas-Benitez, E, Rudino-Pinera, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Preserving metallic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase
To be Published
|
|
8TWM
 
 | |
5K4V
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ refined to 2.2 angstroms | | Descriptor: | ACETATE ION, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UIO
 
 | |
5K7A
 
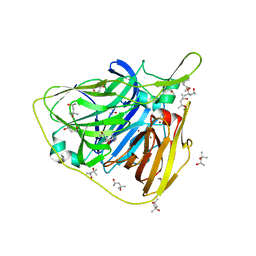 | | Crystal structure of laccase fron Thermus thermophilus HB27 (sodium nitrate 1.5 min) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Laccase | | Authors: | Diaz-Vilchis, A, Ruiz-Arellano, R.R, Rosas-Benitez, E, Rudino-Pinera, E. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Preserving metalic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase
To be Published
|
|
4Y8S
 
 | |
5UM9
 
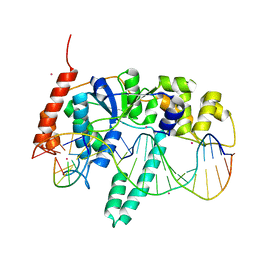 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
6U96
 
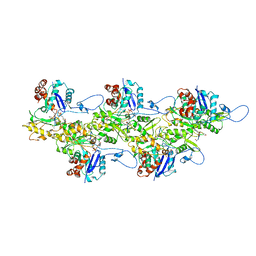 | | Actin phalloidin at BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Das, S, Ge, P, Durer, Z.A.O, Grintsevich, E.E, Zhou, Z.H, Reisler, E. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | D-loop Dynamics and Near-Atomic-Resolution Cryo-EM Structure of Phalloidin-Bound F-Actin.
Structure, 28, 2020
|
|
4Y8I
 
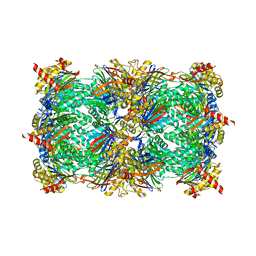 | | Yeast 20S proteasome in complex with Ac-PLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4YA9
 
 | |
6NY2
 
 | | CasX-gRNA-DNA(45bp) state I | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
5JQX
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with phosphoglyceric acid (PGA) - GpgS*PGA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
6O1L
 
 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression Hfq-Crc-amiE 2:3:2 complex | | Descriptor: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | Authors: | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | Deposit date: | 2019-02-20 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
8UWV
 
 | |
6UEU
 
 | |
6NJ9
 
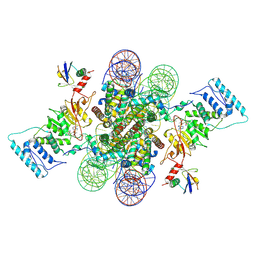 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 2-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-02 | | Release date: | 2019-02-20 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
