5WWU
 
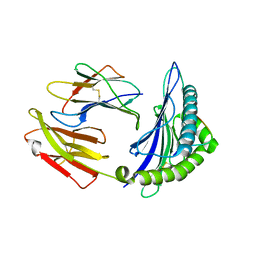 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5WER
 
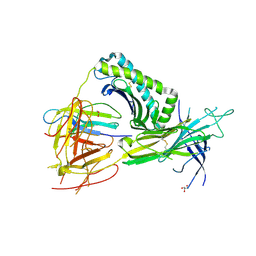 | | Crystal Structure of TAPBPR and H2-Dd complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
4HM0
 
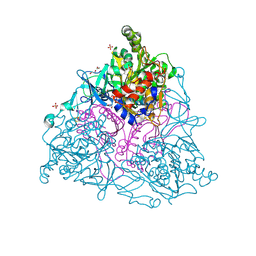 | | Naphthalene 1,2-Dioxygenase bound to indole-3-acetate | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
5WFM
 
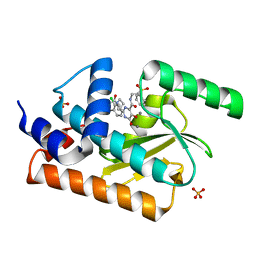 | | Crystal structure of the influenza virus PA endonuclease (E119D mutant) in complex with inhibitor 10e (SRI-30024) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-[2-(phenylsulfonyl)ethyl]-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-07-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W75
 
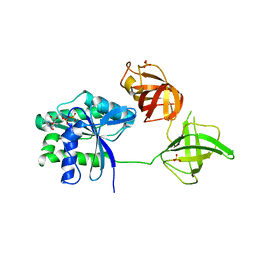 | |
5WFQ
 
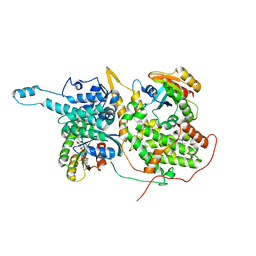 | | Ligand-bound Ras:SOS:Ras complex | | Descriptor: | 7-chloranyl-~{N}-(3-chloranyl-4-fluoranyl-phenyl)-1,2,3,4-tetrahydroacridin-9-amine, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Sun, Q, Phan, J, Burns, M.C, Fesik, S.W. | | Deposit date: | 2017-07-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling.
Anal. Biochem., 548, 2018
|
|
5WFW
 
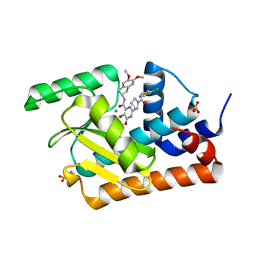 | | Crystal structure of the influenza virus PA endonuclease (E119D mutant) in complex with inhibitor 10j (SRI-30026) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-N-(5,6-dimethoxy-2,3-dihydro-1H-inden-2-yl)-5-hydroxy-6-ox o-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-07-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
4HPS
 
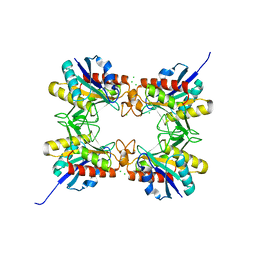 | |
3NH6
 
 | | Nucleotide Binding Domain of human ABCB6 (apo structure) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, BETA-MERCAPTOETHANOL | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5W7O
 
 | |
5WIP
 
 | | TraE protein in complex with 2-(2-furyl)isonicotinic acid | | Descriptor: | 2-(furan-2-yl)pyridine-4-carboxylic acid, Conjugal transfer protein | | Authors: | Casu, B, Arya, T, Bessette, B, Baron, C. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Fragment-based screening identifies novel targets for inhibitors of conjugative transfer of antimicrobial resistance by plasmid pKM101.
Sci Rep, 7, 2017
|
|
3NHF
 
 | |
5WG6
 
 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5WHA
 
 | | KRas G12V, bound to GDP and miniprotein 225-11 | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-16 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
3NHR
 
 | |
5WJ2
 
 | |
5WHG
 
 | | Vms1 mitochondrial localization core | | Descriptor: | Protein VMS1, ZINC ION | | Authors: | Fredrickson, E.K, Schubert, H.L, Rutter, J, Hill, C.P. | | Deposit date: | 2017-07-17 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sterol Oxidation Mediates Stress-Responsive Vms1 Translocation to Mitochondria.
Mol. Cell, 68, 2017
|
|
4HO5
 
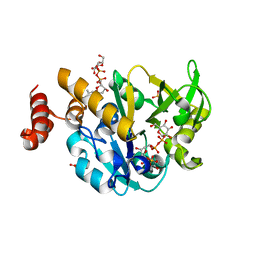 | |
5WJB
 
 | | Crystal Structure of Amino Acids 1733-1797 of Human Beta Cardiac Myosin Fused to Gp7 | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5WHK
 
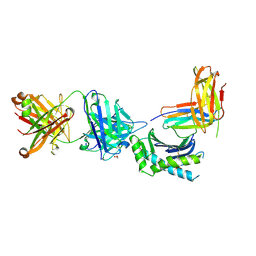 | | Crystal structure of Fab fragment of antibody DX-2507 bound to FcRn-B2M | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Edwards, T.E, Clifton, M.C, Nixon, A.E, Kenniston, J.A. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
J. Biol. Chem., 292, 2017
|
|
5WJP
 
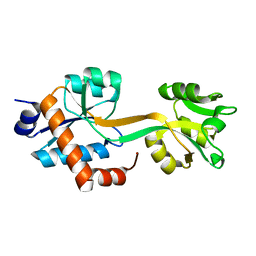 | |
3EEK
 
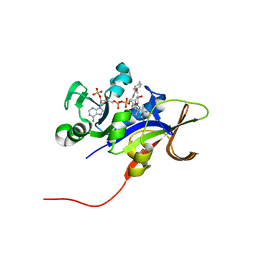 | |
5WHS
 
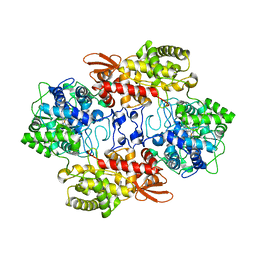 | | Crystal structure of the catalase-peroxidase from Neurospora crassa at 2.6 A | | Descriptor: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz-Vilchis, A, Vega-Garcia, V, Rudino-Pinera, E, Hansberg, W. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure, kinetics, molecular and redox properties of a cytosolic and developmentally regulated fungal catalase-peroxidase.
Arch. Biochem. Biophys., 640, 2018
|
|
3NIJ
 
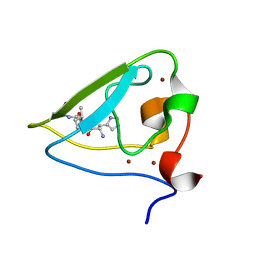 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
5WJV
 
 | |
