4A9F
 
 | |
4ALH
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3,5 dimethyl-4-phenyl-1,2- oxazole | | Descriptor: | 1,2-ETHANEDIOL, 3,5 DIMETHYL-4-PHENYL-1,2-OXAZOLE, BROMODOMAIN CONTAINING 2, ... | | Authors: | Chung, C.W, Bamborough, P. | | Deposit date: | 2012-03-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 2: Optimization of Phenylisoxazole Sulfonamides.
J.Med.Chem., 55, 2012
|
|
4A9J
 
 | |
1V7A
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
3ZWT
 
 | | Structure of Human Dihydroorotate Dehydrogenase with a Bound Inhibitor | | Descriptor: | 2-{[(2,5-DICHLOROPHENYL)METHYL]SULFANYL}-5-ETHYL-[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Acklam, P.A, Parsons, M.R. | | Deposit date: | 2011-08-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Factors Influencing the Specificity of Inhibitor Binding to the Human and Malaria Parasite Dihydroorotate Dehydrogenases.
J.Med.Chem., 55, 2012
|
|
4B78
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (3R,5R)-3-methoxy-5-(4-methoxyphenyl)-5-(3-pyridin-3-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
 | | Crystal structure of MEK1 in complex with fragment 1 | | Descriptor: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZPU
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[3-[(3Z,8S,11R)-8-tert-butyl-11-oxidanyl-7,10-bis(oxidanylidene)-6,9-diazabicyclo[11.2.2]heptadeca-1(15),3,13,16-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors
J.Med.Chem., 56, 2013
|
|
3ZLW
 
 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8Q2J
 
 | | Tau - AD-MIA2 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q27
 
 | | Tau: AD-MIA1 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W, Goedert, M, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q2K
 
 | | Tau - AD-MIA3 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q2L
 
 | | Tau - AD-MIA4 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
4B72
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (6R)-6-(4-methoxyphenyl)-2-methyl-6-(3-pyrimidin-5-ylphenyl)pyrrolo[3,4-d][1,3]thiazol-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
5AR8
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biphenylsulfonamide | | Descriptor: | 2,6-bis(fluoranyl)-N-[3-[5-[2-[(3-methylsulfonylphenyl)amino]pyrimidin-4-yl]-2-morpholin-4-yl-1,3-thiazol-4-yl]phenyl]benzenesulfonamide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
4A7B
 
 | |
4A9E
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3-methyl-1,2,3,4- tetrahydroquinazolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery.
J.Med.Chem., 55, 2012
|
|
4B77
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (5R)-5-(4-methoxyphenyl)-5-(3-pyrimidin-5-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4B70
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-[3-(3-chlorophenyl)phenyl]-2-methyl-5,6-dihydro-1,3-oxazin-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
5AR5
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Benzimidazole | | Descriptor: | 2-(2-(4-CHLOROPHENYL)-1H-IMIDAZOL-5-YL)-N,1-BIS(2-METHOXYETHYL)-1H-BENZO[D]IMIDAZOLE-5-CARBOXAMIDE, CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
5AR4
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with SB-203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
5AR3
 
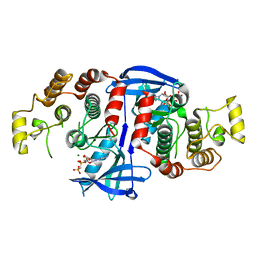 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
3ZWS
 
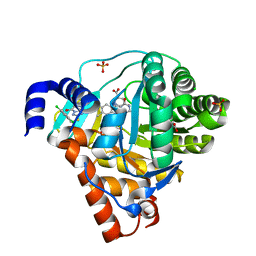 | | Structure of Human Dihydroorotate Dehydrogenase with a Bound Inhibitor | | Descriptor: | 2-[(2,5-DICHLOROBENZYL)SULFANYL]-5-METHYL[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Acklam, P.A, Parsons, M.R. | | Deposit date: | 2011-08-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Factors Influencing the Specificity of Inhibitor Binding to the Human and Malaria Parasite Dihydroorotate Dehydrogenases.
J.Med.Chem., 55, 2012
|
|
3ZEI
 
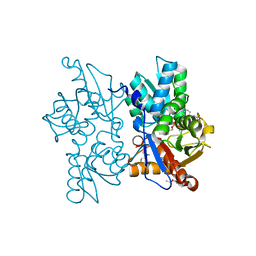 | | Structure of the Mycobacterium tuberculosis O-Acetylserine Sulfhydrylase (OASS) CysK1 in complex with a small molecule inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(Z)-[(5Z)-5-[[2-(2-hydroxy-2-oxoethyloxy)phenyl]methylidene]-3-methyl-4-oxidanylidene-1,3-thiazolidin-2-ylidene]amino]benzoic acid, O-ACETYLSERINE SULFHYDRYLASE, ... | | Authors: | Poyraz, O, Schnell, R, Schneider, G. | | Deposit date: | 2012-12-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Novel Thiazolidine Inhibitors of O-Acetyl Serine Sulfhydrylase from Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
