1IL1
 
 | | Crystal structure of G3-519, an anti-HIV monoclonal antibody | | Descriptor: | monoclonal antibody G3-519 (heavy chain), monoclonal antibody G3-519 (light chain) | | Authors: | Berry, M.B, Johnson, K.A, Radding, W, Fung, M, Liou, R, Phillips Jr, G.N. | | Deposit date: | 2001-05-07 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an anti-HIV monoclonal Fab antibody fragment specific to a gp120 C-4 region peptide.
Proteins, 45, 2001
|
|
1IL2
 
 | | Crystal Structure of the E. coli Aspartyl-tRNA Synthetase:Yeast tRNAasp:aspartyl-Adenylate Complex | | Descriptor: | ASPARTYL TRANSFER RNA, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Moulinier, L, Eiler, S, Eriani, G, Gangloff, J, Thierry, J.C, Gabriel, K, McClain, W.H, Moras, D. | | Deposit date: | 2001-05-07 | | Release date: | 2001-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism.
EMBO J., 20, 2001
|
|
1IL3
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 7-DEAZAGUANINE | | Descriptor: | 7-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL4
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 9-DEAZAGUANINE | | Descriptor: | 9-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL5
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 2,5-DIAMINO-4,6-DIHYDROXYPYRIMIDINE (DDP) | | Descriptor: | 2,4-DIAMINO-4,6-DIHYDROXYPYRIMIDINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL6
 
 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
1IL8
 
 | |
1IL9
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 8-METHYL-9-OXOGUANINE | | Descriptor: | 5-AMINO-2-METHYL-6H-OXAZOLO[5,4-D]PYRIMIDIN-7-ONE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1ILC
 
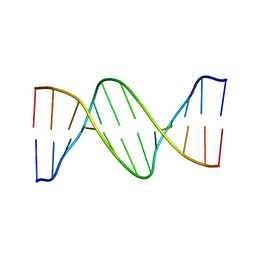 | | DNA Bending by an Adenine-Thymine Tract and Its Role in Gene Regulation. | | Descriptor: | 5'-D(*AP*CP*CP*GP*AP*AP*TP*TP*CP*GP*GP*T)-3' | | Authors: | Hizver, J, Rozenberg, H, Frolow, F, Rabinovich, D, Shakked, Z. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA bending by an adenine--thymine tract and its role in gene regulation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ILD
 
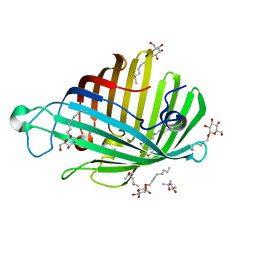 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1ILE
 
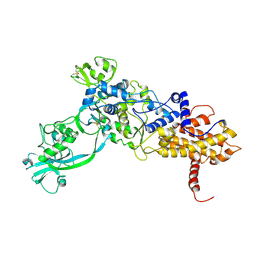 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
1ILF
 
 | |
1ILG
 
 | |
1ILH
 
 | |
1ILK
 
 | |
1ILO
 
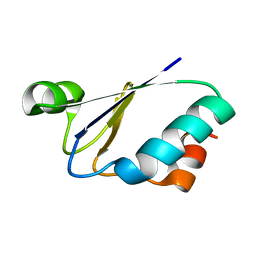 | | NMR structure of a thioredoxin, MtH895, from the archeon Methanobacterium thermoautotrophicum strain delta H. | | Descriptor: | conserved hypothetical protein MtH895 | | Authors: | Bhattacharyya, S, Habibi-Nazhad, B, Slupsky, C.M, Sykes, B.D, Wishart, D.S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification of a novel archaebacterial thioredoxin: determination of function through structure.
Biochemistry, 41, 2002
|
|
1ILP
 
 | |
1ILQ
 
 | |
1ILR
 
 | |
1ILS
 
 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
1ILT
 
 | |
1ILU
 
 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
1ILW
 
 | |
1ILX
 
 | | Excited State Dynamics in Photosystem II Revised. New Insights from the X-ray Structure. | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Vasilev, S, Orth, P, Zouni, A, Owens, T.G, Bruce, D. | | Deposit date: | 2001-05-09 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Excited-state dynamics in photosystem II: insights from the x-ray crystal structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
