5OLX
 
 | |
5OJ7
 
 | |
5OK1
 
 | |
5XGF
 
 | |
5OLS
 
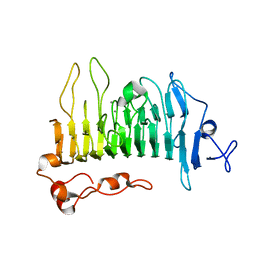 | | Rhamnogalacturonan lyase | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5XDP
 
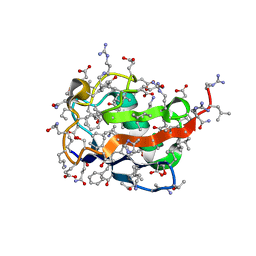 | |
5XPJ
 
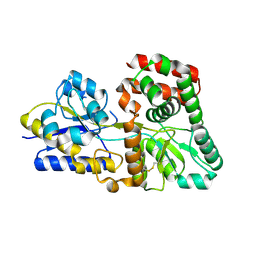 | |
3WFW
 
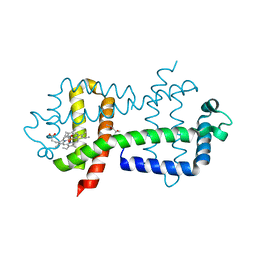 | | Crystal Structure of the Closed Form of the HGbRL's Globin Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hemoglobin-like flavoprotein fused to Roadblock/LC7 domain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teh, A.H. | | Deposit date: | 2013-07-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Open and Lys-His Hexacoordinated Closed Structures of a Globin with Swapped Proximal and Distal Sites.
Sci Rep, 5, 2015
|
|
3WNR
 
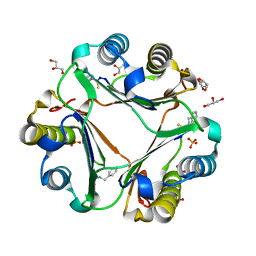 | | Multiple binding modes of benzyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
5XCN
 
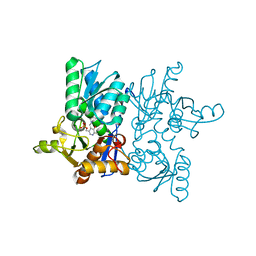 | |
5XRF
 
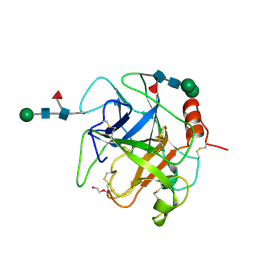 | | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Snake venom serine protease Da-36, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, C.-C, Wu, W.-G, Fan, Q, Tian, J, Zhang, Z, Zheng, Y. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus
To Be Published
|
|
4EHW
 
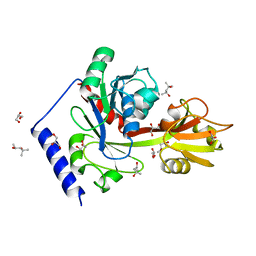 | | Crystal structure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5X3M
 
 | |
5XAO
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala in complexes with sodium and chloride ions | | Descriptor: | ACETIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kamitori, S, Sode, K. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
5XC6
 
 | | Dengue Virus 4 NS3 Helicase in complex with SSRNA SLA12 | | Descriptor: | NS3 Helicase, PHOSPHATE ION, RNA (5'-R(*AP*GP*UP*UP*GP*UP*UP*AP*GP*UP*CP*U)-3') | | Authors: | Swarbrick, C.M.D, Basavannacharya, C, Chan, K.W.K, Chan, S.A, Singh, D, Wei, N, Phoo, W.W, Luo, D, Lescar, J, Vasudevan, S.G. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication
Nucleic Acids Res., 45, 2017
|
|
5XOQ
 
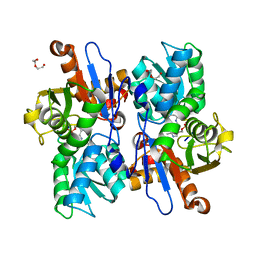 | |
5XPK
 
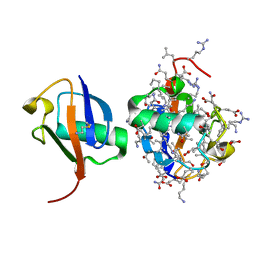 | |
4C61
 
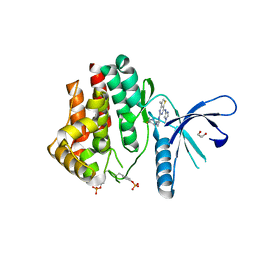 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | ACETATE ION, N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-7-methyl-N4-(1-methylimidazol-4-yl)thieno[3,2-d]pyrimidine-2,4-diamine, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J.A, Green, I, Pollard, H, Howard, T. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 1-Methyl-1H-Imidazole Derivatives as Potent Jak2 Inhibitors.
J.Med.Chem., 57, 2014
|
|
5XPX
 
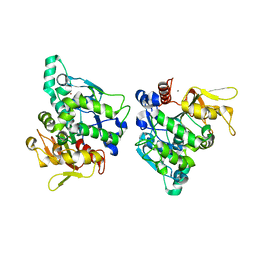 | |
4DH3
 
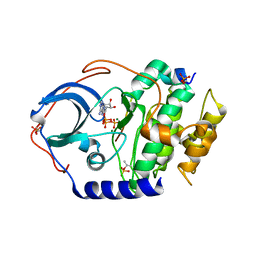 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7Y37
 
 | |
4DIK
 
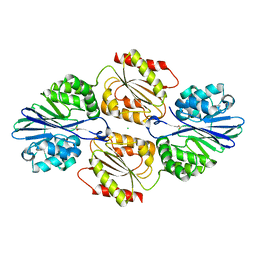 | | Flavo Di-iron protein H90A mutant from Thermotoga maritima | | Descriptor: | CHLORIDE ION, FLAVOPROTEIN, MU-OXO-DIIRON | | Authors: | Fang, H, Caranto, J.D, Taylor, A.B, Hart, P.J, Kurtz Jr, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Histidine ligand variants of a flavo-diiron protein: effects on structure and activities.
J.Biol.Inorg.Chem., 17, 2012
|
|
4DN4
 
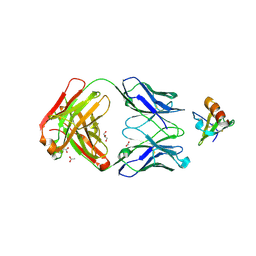 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
5XC7
 
 | | Dengue Virus 4 NS3 Helicase D290A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 Helicase | | Authors: | Swarbrick, C.M.D, Basavannacharya, C, Chan, K.W.K, Chan, S.A, Singh, D, Wei, N, Phoo, W.W, Luo, D, Lescar, J, Vasudevan, S.G. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication
Nucleic Acids Res., 45, 2017
|
|
