5OH2
 
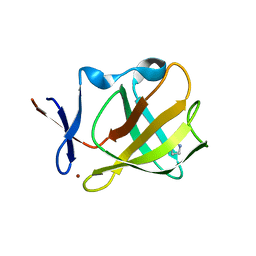 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OHB
 
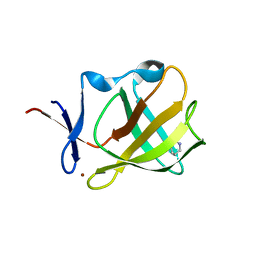 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidin-2-one (Valerolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
3DBK
 
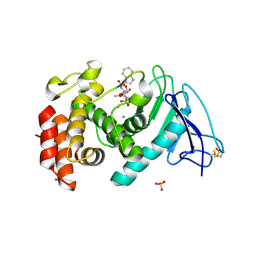 | | Pseudomonas aeruginosa elastase with phosphoramidon | | Descriptor: | CALCIUM ION, Elastase, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | McKay, D.B, Overgaard, M.T. | | Deposit date: | 2008-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Elastase of Pseudomonas aeruginosa Complexed with Phosphoramidon
To be Published
|
|
1O9O
 
 | |
4PEX
 
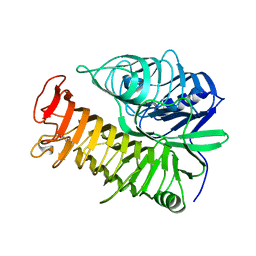 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
3D9K
 
 | |
7B4J
 
 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
7AFZ
 
 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
3VS7
 
 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Honda, K, Niwa, H, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
1NY6
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the active state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, transcriptional regulator (NtrC family) | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
5OH1
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Aminoglutethimide | | Descriptor: | (3~{R})-3-(4-aminophenyl)-3-ethyl-piperidine-2,6-dione, Cereblon isoform 4, S-Thalidomide, ... | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH9
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thiazolidine-2,4-dione | | Descriptor: | 1,3-thiazole-2,4-dione, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH3
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Ethosuximide | | Descriptor: | (3~{S})-3-ethyl-3-methyl-pyrrolidine-2,5-dione, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5AC4
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
3D9O
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
5OHA
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with 2-Thiohydantoin | | Descriptor: | 2-sulfanylideneimidazol-4-one, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Imidazolidine-2,4-dione (Hydantoin) | | Descriptor: | Cereblon isoform 4, ZINC ION, imidazolidine-2,4-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
3VFZ
 
 | |
4GS8
 
 | | Structure analysis of cysteine free insulin degrading enzyme (ide) with compound bdm43079 [{[(s)-2-(1h-imidazol-4-yl)-1-methylcarbamoyl-ethylcarbamoyl]-methyl}-(3-phenyl-propyl)-amino]-acetic acid | | Descriptor: | Insulin-degrading enzyme, N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-N-methyl-L-histidinamide, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
5A5G
 
 | | Crystal structure of FTHFS2 from T.acetoxydans Re1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMATE--TETRAHYDROFOLATE LIGASE, GLYCEROL, ... | | Authors: | Bergdahl, R, Jacobson, F, Muller, B, Mikkelsen, N, Schurer, A, Sandgren, M. | | Deposit date: | 2015-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization, Crystallization and Three- Dimensional Structures of Formyltetrahydrofolate Synthetase (Fthfs) from the Syntrophic Acetate Oxidising Bacterium Tepidanaerobacter Acetatoxydans Re1
To be Published
|
|
4GWS
 
 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase with Filled Central Cavity | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Gao, Y, Honzatko, R.B. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hydrophobic Central Cavity in Fructose-1,6-bisphosphatase is Essential for the Synergism in AMP/Fructose 2,6-bisphosphate Inhibition
To be Published
|
|
4GYJ
 
 | |
7SLN
 
 | | [U:Ag+:U--pH 10.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7SLJ
 
 | | [U:Ag+:U--pH 8.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
