1EN6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146L MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
2BQZ
 
 | | Crystal structure of a ternary complex of the human histone methyltransferase Pr-SET7 (also known as SET8) | | Descriptor: | HISTONE H4, S-ADENOSYL-L-HOMOCYSTEINE, SET8 PROTEIN | | Authors: | Xiao, B, Jing, C, Kelly, G, Walker, P.A, Muskett, F.W, Frenkiel, T.A, Martin, S.R, Sarma, K, Reinberg, D, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Genes Dev., 19, 2005
|
|
1EEF
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH BOUND LIGAND PEPG | | Descriptor: | 2-PHENETHYL-2,3-DIHYDRO-PHTHALAZINE-1,4-DIONE, PROTEIN (HEAT-LABILE ENTEROTOXIN B CHAIN), alpha-D-galactopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8HXN
 
 | | Crystal structure of B2 Sfh-I MBL in complex with 2-amino-5-(4-(but-3-en-1-yloxy)benzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-but-3-enoxyphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Beta-lactamase, ETHANOL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
2BGM
 
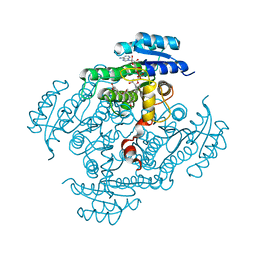 | | X-Ray structure of ternary-Secoisolariciresinol Dehydrogenase | | Descriptor: | MATAIRESINOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
5HW1
 
 | |
2BTO
 
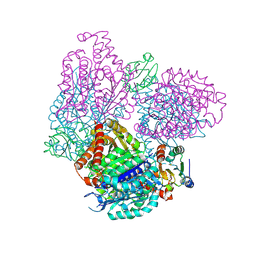 | | Structure of BtubA from Prosthecobacter dejongeii | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, THIOREDOXIN 1, TUBULIN BTUBA | | Authors: | Schlieper, D, Lowe, J. | | Deposit date: | 2005-06-04 | | Release date: | 2005-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Bacterial Tubulin Btuba/B: Evidence for Horizontal Gene Transfer.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4PJH
 
 | |
2RIC
 
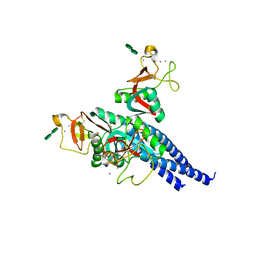 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptopyranosyl-(1-3)-L-glycero-D-manno-heptopyranose | | Descriptor: | CALCIUM ION, L-glycero-alpha-D-manno-heptopyranose, L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose, ... | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
2BOZ
 
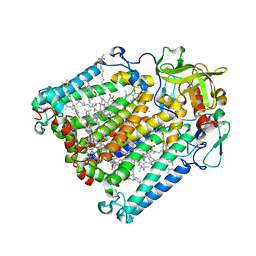 | | Photosynthetic Reaction Center Mutant With Gly M203 Replaced With Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, DECANE, ... | | Authors: | Potter, J.P, Fyfe, P.K, Jones, M.R. | | Deposit date: | 2005-04-15 | | Release date: | 2005-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Strong Effects of an Individual Water Molecule on the Rate of Light-Driven Charge Separation in the Rhodobacter Sphaeroides Reaction Center.
J.Biol.Chem., 280, 2005
|
|
9ASS
 
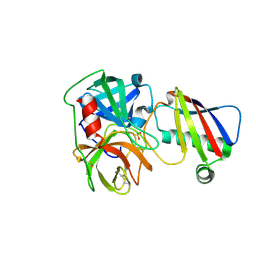 | |
1EC6
 
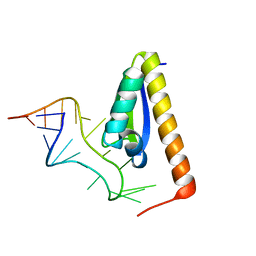 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN BOUND TO 20-MER RNA HAIRPIN | | Descriptor: | 20-MER RNA HAIRPIN, RNA-BINDING PROTEIN NOVA-2 | | Authors: | Lewis, H.A, Musunuru, K, Jensen, K.B, Edo, C, Chen, H. | | Deposit date: | 2000-01-25 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome.
Cell(Cambridge,Mass.), 100, 2000
|
|
1EDE
 
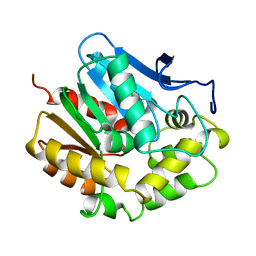 | |
7ALG
 
 | | The RSLex - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
1EN5
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Y34F MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
5I5C
 
 | | X-ray crystal structure of allo-Thr31-ShK | | Descriptor: | GLYCEROL, Kappa-stichotoxin-She3a, LITHIUM ION, ... | | Authors: | Dang, B, Kubota, T, Manda, K.l, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1NWS
 
 | | Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3 like protein 1 | | Authors: | Fusetti, F, Pijning, T, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2003-02-06 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39
J.Biol.Chem., 278, 2003
|
|
2RIA
 
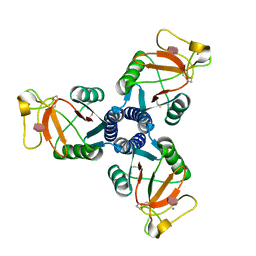 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with D-glycero-D-manno-heptose | | Descriptor: | CALCIUM ION, D-glycero-alpha-D-manno-heptopyranose, Pulmonary surfactant-associated protein D | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
1EN4
 
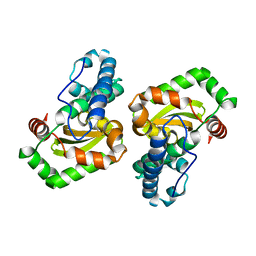 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146H MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EBF
 
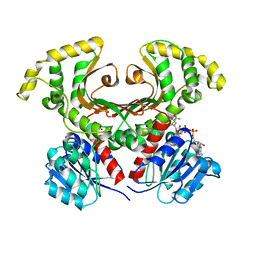 | | HOMOSERINE DEHYDROGENASE FROM S. CEREVISIAE COMPLEX WITH NAD+ | | Descriptor: | HOMOSERINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
5O0N
 
 | | Deglycosylated Nogo Receptor with native disulfide structure 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Reticulon-4 receptor, ... | | Authors: | Pronker, M.F, Tas, R.P, Vlieg, H.C, Janssen, B.J.C. | | Deposit date: | 2017-05-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nogo Receptor crystal structures with a native disulfide pattern suggest a novel mode of self-interaction.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2C68
 
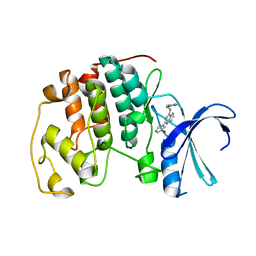 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDROPYRAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2BYR
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA in complex with methyllycaconitine | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, METHYLLYCACONITINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
