6DBU
 
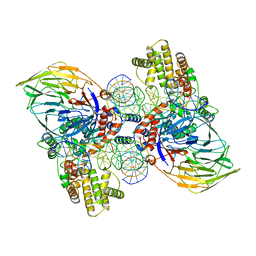 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
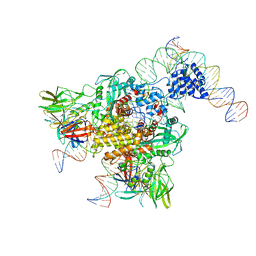 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1JVX
 
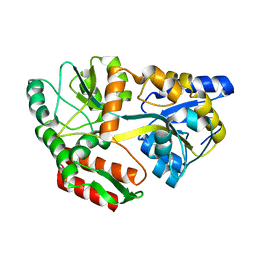 | | Maltodextrin-binding protein variant D207C/A301GS/P316C cross-linked in crystal | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltodextrin-binding protein | | Authors: | Srinivasan, U, Iyer, G.H, Przybycien, T.A, Samsonoff, W.A, Bell, J.A. | | Deposit date: | 2001-08-31 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystine: Fibrous Biomolecular Material from Protein Crystals Cross-linked in a Specific Geometry
Protein Eng., 15, 2002
|
|
1JW5
 
 | |
6DBJ
 
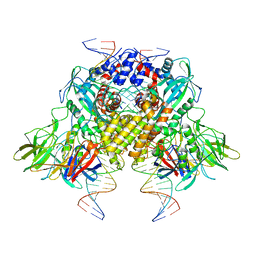 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DKS
 
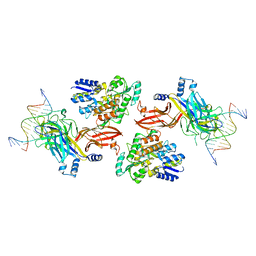 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
6D67
 
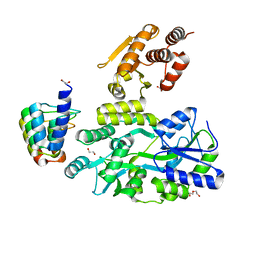 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion (maltose bound form) in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Designed AR protein mbp3_16, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DBO
 
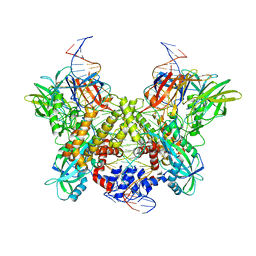 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1LAX
 
 | | CRYSTAL STRUCTURE OF MALE31, A DEFECTIVE FOLDING MUTANT OF MALTOSE-BINDING PROTEIN | | Descriptor: | MALTOSE-BINDING PROTEIN MUTANT MALE31, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Mourez, M, Vulliez-le Normand, B, Sassoon, N, Bentley, G.A, Betton, J.M. | | Deposit date: | 2002-03-29 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a defective folding protein
PROTEIN SCI., 12, 2003
|
|
5FIO
 
 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
5GS2
 
 | | Crystal structure of diabody complex with repebody and MBP | | Descriptor: | Maltose-binding periplasmic protein, anti-MBP, anti-repebody, ... | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
6EQZ
 
 | | A MamC-MIC insertion in MBP scaffold at position K170 | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Zarivach, R. | | Deposit date: | 2017-10-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1LLS
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
6EG3
 
 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
5GPP
 
 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5GRU
 
 | | Structure of mono-specific diabody | | Descriptor: | Maltose-binding periplasmic protein, diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GXT
 
 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
5GPQ
 
 | | Crystal Structure of zebrafish ASC CARD Domain | | Descriptor: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5FSG
 
 | |
1MH3
 
 | | maltose binding-a1 homeodomain protein chimera, crystal form I | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
5EDU
 
 | |
1MH4
 
 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
5GXV
 
 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
