1QB6
 
 | | BOVINE TRYPSIN 3,3'-[3,5-DIFLUORO-4-METHYL-2, 6-PYRIDINEDIYLBIS(OXY)]BIS(BENZENECARBOXIMIDAMIDE) (ZK-805623) COMPLEX | | Descriptor: | 3,3'-[3,5-DIFLUORO-4-METHYL-2,6-PYRIDYLENEBIS(OXY)]-BIS(BENZENECARBOXIMIDAMIDE), CALCIUM ION, POTASSIUM ION, ... | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QBO
 
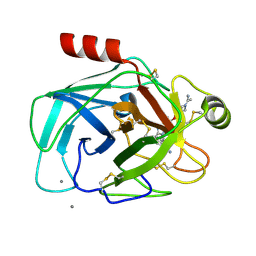 | |
1QB1
 
 | |
4QDO
 
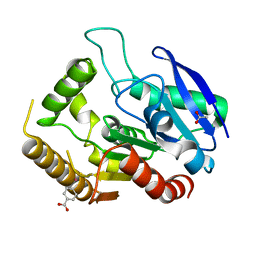 | |
1LSU
 
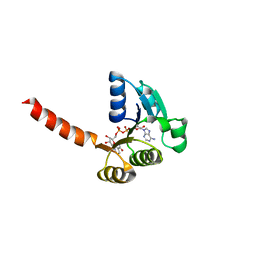 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
4QDU
 
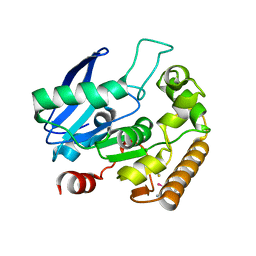 | |
4QE3
 
 | | Crystal structure of Antigen 85C-H260Q mutant | | Descriptor: | ACETIC ACID, CHAPSO, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QNV
 
 | | Crystal structure of Cx-SAM bound CmoB from E. coli in P6122 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4QNZ
 
 | |
1QLP
 
 | | 2.0 ANGSTROM STRUCTURE OF INTACT ALPHA-1-ANTITRYPSIN: A CANONICAL TEMPLATE FOR ACTIVE SERPINS | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Elliott, P.R, Pei, X.Y, Dafforn, T, Read, R.J, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1999-09-10 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease.
Protein Sci., 9, 2000
|
|
1QLZ
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ECZ
 
 | | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Saito, K, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1
To be Published
|
|
3JZU
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ELY
 
 | |
4QO0
 
 | |
4QN1
 
 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
1QZM
 
 | | alpha-domain of ATPase | | Descriptor: | ATP-dependent protease La | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
2E7L
 
 | |
3ZI8
 
 | | Structure of the R17A mutant of the Ralstonia soleanacerum lectin at 1.5 Angstrom in complex with L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MAGNESIUM ION, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Arnaud, J, Audfray, A, Claudinon, J, Trondle, K, Trosvalet, M, Thomas, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reduction of Lectin Valency Drastically Changes Glycolipid Dynamics in Membranes, But not Surface Avidity.
Acs Chem.Biol., 8, 2013
|
|
2E1P
 
 | | Crystal structure of pro-Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Saito, K, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of unautoprocessed precursor of subtilisin from a hyperthermophilic archaeon: evidence for Ca2+-induced folding
J.Biol.Chem., 282, 2007
|
|
3X2R
 
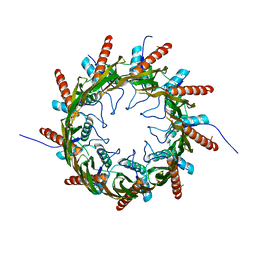 | | Structure of the nonameric bacterial amyloid secretion channel CsgG | | Descriptor: | CsgG | | Authors: | Huang, Y, Cao, B, Zhao, Y, Kou, Y, Ni, D, Zhang, X.C. | | Deposit date: | 2014-12-29 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the nonameric bacterial amyloid secretion channel
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1LEK
 
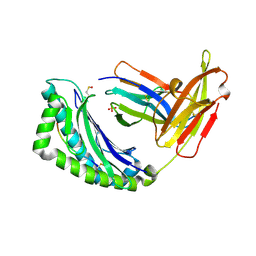 | | Crystal Structure of H-2Kbm3 bound to dEV8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Luz, J.G, Huang, M, Garcia, K.C, Rudolph, M.G, Apostolopoulos, V, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-04-09 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
J.Exp.Med., 195, 2002
|
|
1NO8
 
 | | SOLUTION STRUCTURE OF THE NUCLEAR FACTOR ALY RBD DOMAIN | | Descriptor: | ALY | | Authors: | Perez-Alvarado, G.C, Martinez-Yamout, M, Allen, M.M, Grosschedl, R, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-01-15 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Nuclear Factor ALY: Insights into Post-Transcriptional
Regulatory and mRNA Nuclear Export Processes
Biochemistry, 42, 2003
|
|
1LUI
 
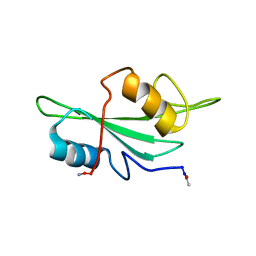 | | NMR Structures of Itk SH2 domain, Pro287cis isoform, ensemble of 20 low energy structures | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, B.F, Andreotti, A.M. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
