3ZWI
 
 | | RECOMBINANT NATIVE CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND AT 1.25 A:UNRESTRAINT REFINEMENT | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-07-31 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas-Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4R7X
 
 | | Crystal structure of N-lobe of human ARRDC3(1-180) | | Descriptor: | Arrestin domain-containing protein 3, PHOSPHATE ION | | Authors: | Qi, S, Hurley, J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Insights into beta 2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3.
Protein Sci., 23, 2014
|
|
4TVG
 
 | | HIV Protease (PR) dimer in closed form with pepstatin in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, GLYCEROL, HIV Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
3GVP
 
 | | Human SAHH-like domain of human adenosylhomocysteinase 3 | | Descriptor: | Adenosylhomocysteinase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Siponen, M.I, Wisniewska, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schueler, H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human S-adenosyl homocysteine hydrolase-like 2 protein crystal structure
To be Published
|
|
3GG6
 
 | | Crystal structure of the NUDIX domain of human NUDT18 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 18 | | Authors: | Tresaugues, L, Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NUDIX domain of human NUDT18
To be Published
|
|
3ZQV
 
 | |
4TVH
 
 | | HIV Protease (PR) dimer in closed form with TL-3 in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, BETA-MERCAPTOETHANOL, Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
3H95
 
 | | Crystal structure of the NUDIX domain of NUDT6 | | Descriptor: | CITRATE ANION, GLYCEROL, Nucleoside diphosphate-linked moiety X motif 6 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotyenova, T, Kotzch, A, Nielsen, T.K, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the NUDIX domain of NUDT6
To be Published
|
|
3DH1
 
 | | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Welin, M, Tresaugues, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, van der Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2.
To be Published
|
|
3EC8
 
 | | The crystal structure of the RA domain of FLJ10324 (RADIL) | | Descriptor: | CHLORIDE ION, GLYCEROL, LEAD (II) ION, ... | | Authors: | Wisniewska, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the RA domain of FLJ10324 (RADIL)
to be published
|
|
5AP9
 
 | | Controlled lid-opening in Thermomyces lanuginosus lipase - a switch for activity and binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Skjold-Joergensen, J, Vind, J, Moroz, O.V, Blagova, E.V, Bhatia, V.K, Svendsen, A, Wilson, K.S, Bjerrum, M.J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function.
Biochim. Biophys. Acta, 1865, 2017
|
|
1F09
 
 | | CRYSTAL STRUCTURE OF THE GREEN FLUORESCENT PROTEIN (GFP) VARIANT YFP-H148Q WITH TWO BOUND IODIDES | | Descriptor: | GREEN FLUORESCENT PROTEIN, IODIDE ION | | Authors: | Wachter, R.M, Yarbrough, D, Kallio, K, Remington, S.J. | | Deposit date: | 2000-05-15 | | Release date: | 2000-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein.
J.Mol.Biol., 301, 2000
|
|
7T4D
 
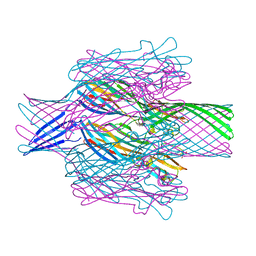 | | Pore structure of pore-forming toxin Epx4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | Authors: | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
1F0B
 
 | |
3EWS
 
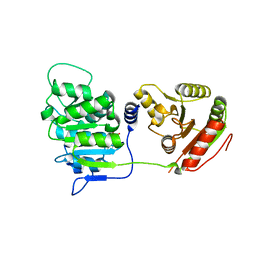 | | Human DEAD-box RNA-helicase DDX19 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B | | Authors: | Lehtio, l, Karlberg, t, Andersson, j, Arrowsmith, c.h, Berglund, h, Bountra, c, Collins, r, Dahlgren, l.g, Edwards, a.m, Flodin, s, Flores, a, Graslund, s, Hammarstrom, m, Johansson, a, Johansson, i, Kotenyova, t, Moche, m, Nilsson, m.e, Nordlund, p, Nyman, t, Olesen, k, Persson, c, Sagemark, j, Thorsell, a.g, Tresaugues, l, Van den berg, s, Weigelt, j, Welin, m, Wikstrom, m, Wisniewska, m, Schueler, h, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
4XK8
 
 | | Crystal structure of plant photosystem I-LHCI super-complex at 2.8 angstrom resolution | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Suga, M, Qin, X, Kuang, T, Shen, J.R. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex
Science, 348, 2015
|
|
8TN1
 
 | |
8TN6
 
 | |
8TNC
 
 | |
8TND
 
 | |
8TNB
 
 | |
3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
5CBX
 
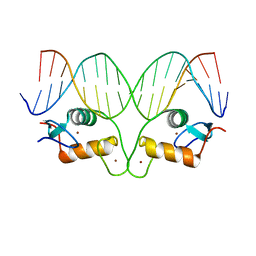 | | AncGR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CC1
 
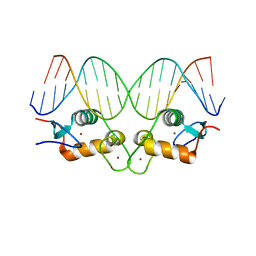 | | S425G Glucocorticoid receptor DNA binding domain - (+)GRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Weikum, E.A, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2JRQ
 
 | | NMR solution structure of the anticodon of E. coli TRNA-VAL3 with 1 modification (cmo5U34) | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*(CM0)P*AP*CP*AP*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Wobble-Position Modifications Pre-structure tRNA's Anticodon for Ribosome-Mediated Codon Binding
To be Published
|
|
