3UV0
 
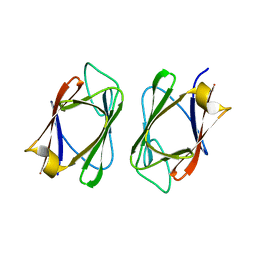 | | Crystal structure of the drosophila MU2 FHA domain | | Descriptor: | Mutator 2, isoform B | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimerization, but not phosphothreonine binding, is conserved between the forkhead-associated domains of Drosophila MU2 and human MDC1
Febs Lett., 586, 2012
|
|
1P9Q
 
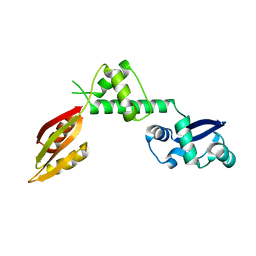 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
2J2I
 
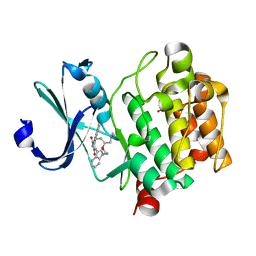 | | Crystal Structure of the humab PIM1 in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1, SULFATE ION | | Authors: | Debreczeni, J.E, Bullock, A.N, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2KE6
 
 | |
2KUR
 
 | |
2KUV
 
 | |
2KXK
 
 | | Human Insulin Mutant A22Gly-B31Lys-B32Arg | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Bocian, W, Kozerski, L. | | Deposit date: | 2010-05-07 | | Release date: | 2011-06-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Novel recombinant insulin analogue with flexible C - terminus in B chain. NMR structure of biosynthetic engineered A22G-B31K-B32R human insulin monomer in water /acetonitrile solution.
To be Published
|
|
2KUW
 
 | |
2C1Y
 
 | | Structure of PDI-related Chaperone, Wind mutant-Y55K | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C0E
 
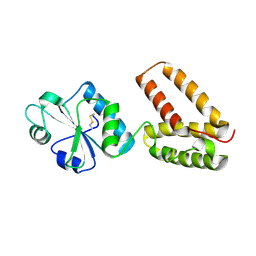 | | Structure of PDI-related Chaperone, Wind with his-tag on C-terminus. | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-01 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1R85
 
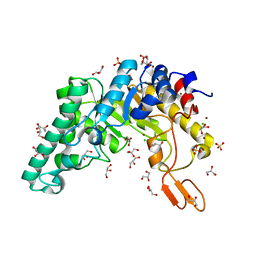 | | Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6): The WT enzyme (monoclinic form) at 1.45A resolution | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Bar, M, Golan, G, Nechama, M, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mapping glycoside hydrolase substrate subsites by isothermal titration calorimetry.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1OIG
 
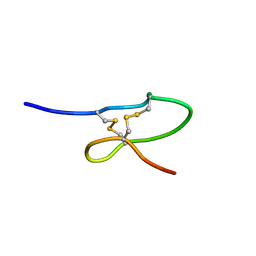 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
1P1C
 
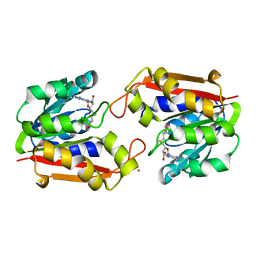 | | Guanidinoacetate Methyltransferase with Gd ion | | Descriptor: | GADOLINIUM ION, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Takusagawa, F. | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Monoclinic guanidinoacetate methyltransferase and gadolinium ion-binding characteristics.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2F7R
 
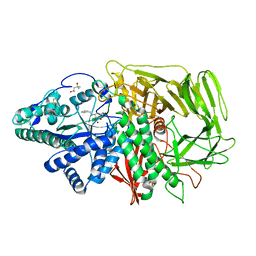 | | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol | | Descriptor: | (1R,2R,3S,4S,5R)-5-(BENZYLAMINO)CYCLOPENTANE-1,2,3,4-TETROL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol
To be Published
|
|
1O2E
 
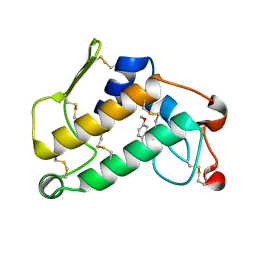 | | Structure of the triple mutant (K53,56,120M) + Anisic acid complex of phospholipase A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the free and anisic acid bound triple mutant of phospholipase A2.
J.Mol.Biol., 333, 2003
|
|
1O3W
 
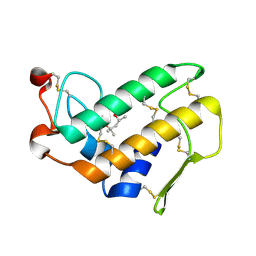 | |
2FNI
 
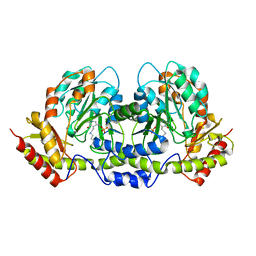 | | PseC aminotransferase involved in pseudoaminic acid biosynthesis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Cygler, M, Matte, A, Lunin, V.V, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FN6
 
 | | Helicobacter pylori PseC, aminotransferase involved in the biosynthesis of pseudoaminic acid | | Descriptor: | AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Cygler, M, Lunin, V.V, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FNU
 
 | | PseC aminotransferase with external aldimine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, aminotransferase | | Authors: | Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
1Q04
 
 | | Crystal structure of FGF-1, S50E/V51N | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1Q03
 
 | | Crystal structure of FGF-1, S50G/V51G mutant | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1PS3
 
 | | Golgi alpha-mannosidase II in complex with kifunensine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase II, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Biochemistry, 42, 2003
|
|
1R33
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
2F7O
 
 | | Golgi alpha-mannosidase II complex with mannostatin A | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis of the Inhibition of Golgi alpha-Mannosidase II by Mannostatin A and the Role of the Thiomethyl Moiety in Ligand-Protein Interactions.
J.Am.Chem.Soc., 128, 2006
|
|
1R34
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamidinium salt | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1Z)-2-phenylethanimidoyl]-5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
