9E0O
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei L-hydrazino-Lysine analog at 2.04 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
6YD5
 
 | | SaFtsZ-UCM151 (comp. 18) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6B3B
 
 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9GQE
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(2,4)-H2 | | Descriptor: | (2~{S},9~{S})-2-cyclohexyl-23,24-dimethoxy-11,17,21-trioxa-4-azatricyclo[20.3.1.0^{4,9}]hexacosa-1(26),22,24-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQD
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(2,4)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-23,24-dimethoxy-11,17,21-trioxa-4-azatricyclo[20.3.1.0^{4,9}]hexacosa-1(26),14,22,24-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQH
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(10,8)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{R},14~{E},18~{R})-2-cyclohexyl-18-(hydroxymethyl)-22,23-dimethoxy-12,13-dimethyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQG
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(10,7)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{R},14~{E},18~{S})-2-cyclohexyl-18-(hydroxymethyl)-22,23-dimethoxy-12,13-dimethyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8PT5
 
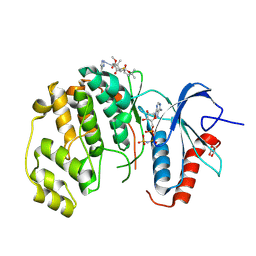 | | ERK2 covelently bound to RU187 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
9GQF
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(2,3)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-22,23-dimethoxy-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQK
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(11,5)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{S},14~{E},18~{S})-2-cyclohexyl-22,23-dimethoxy-12,18-dimethyl-13-oxidanyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQJ
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(11,8)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{S},14~{E},18~{R})-2-cyclohexyl-18-(hydroxymethyl)-22,23-dimethoxy-12-methyl-13-oxidanyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8EFN
 
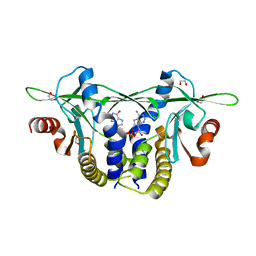 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
9GQI
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(11,6)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{S},14~{E},18~{R})-2-cyclohexyl-22,23-dimethoxy-12,18-dimethyl-13-oxidanyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8BWB
 
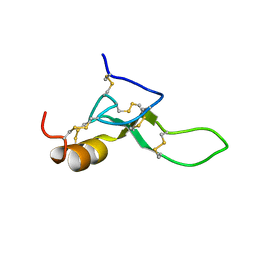 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
9GMR
 
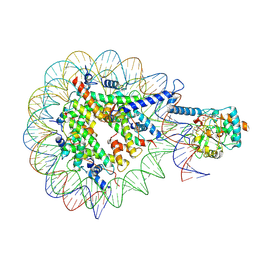 | | SIRT7-H3K36MTUnucleosome complex | | Descriptor: | DNA (149-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
5KEW
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt taurodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tomchick, D.R, Orth, K, Rivera-Cancel, G. | | Deposit date: | 2016-06-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Bile salt receptor complex activates a pathogenic type III secretion system.
Elife, 5, 2016
|
|
3IGG
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFQ with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-[1-(cis-3-hydroxycyclobutyl)-1H-imidazol-4-yl]-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5KFN
 
 | |
8S16
 
 | | c-KIT kinase domain in complex with imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, Mast/stem cell growth factor receptor Kit | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2024-02-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | c-KIT kinase domain in complex with imatinib
To Be Published
|
|
1A95
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH MG:CPRPP AND GUANINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, GUANINE, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
5N1S
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-(1H-Indol-2-yl)-benzenesulfonamide | | Descriptor: | 4-(1~{H}-indol-2-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
4R4C
 
 | | Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)-2-chlorobenzamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({[4-(5-carboxyfuran-2-yl)-2-chlorobenzoyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
7YUG
 
 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
8S18
 
 | | c-KIT kinase domain in complex with S116836 | | Descriptor: | 3-[2-[2-(cyclopropylamino)pyrimidin-5-yl]ethynyl]-~{N}-[3-imidazol-1-yl-5-(trifluoromethyl)phenyl]-4-methyl-benzamide, CHLORIDE ION, Mast/stem cell growth factor receptor Kit | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2024-02-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | c-KIT kinase domain in complex with S116836
To Be Published
|
|
