3F4H
 
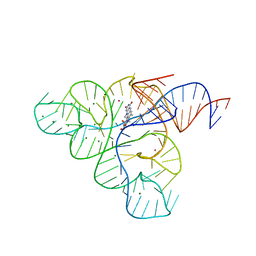 | | Crystal structure of the FMN riboswitch bound to roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, FMN riboswitch, MAGNESIUM ION, ... | | Authors: | Serganov, A.A, Huang, L. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Nature, 458, 2009
|
|
8BT0
 
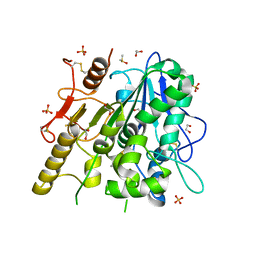 | | Notum Inhibitor ARUK3005518 | | Descriptor: | (3~{a}~{S})-2,2-bis(fluoranyl)-3~{a},4-dihydro-3~{H}-pyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSZ
 
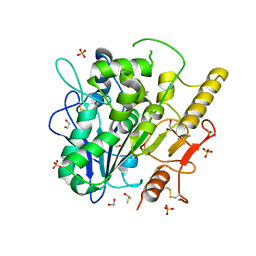 | | Notum Inhibitor ARUK3005522 | | Descriptor: | (3~{a}~{R})-2,3,3~{a},4-tetrahydropyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8G66
 
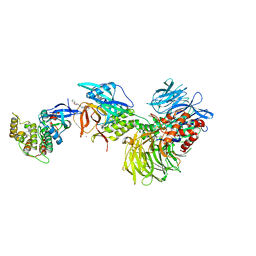 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
8OJ8
 
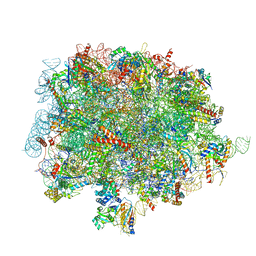 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8V0N
 
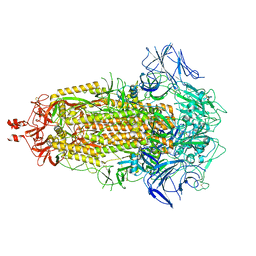 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer 3 (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0Q
 
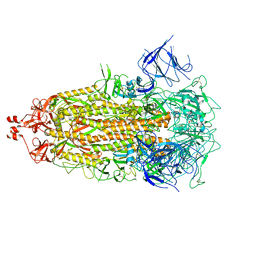 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 3 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0U
 
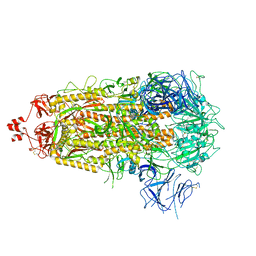 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 4 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
6GJ8
 
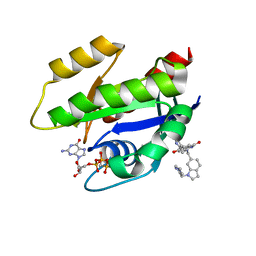 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8BPY
 
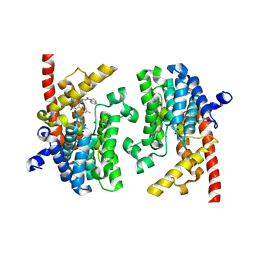 | | X-RAY STRUCTURE OF PDE9A IN COMPLEX WITH Inhibitor 13A | | Descriptor: | (8~{S})-6-[2-(2,3-dihydroindol-1-yl)-2-oxidanylidene-ethyl]-4-(4-methylphenyl)-2-oxidanylidene-8-propyl-1,5,7,8-tetrahydro-1,6-naphthyridine-3-carbonitrile, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Steuber, H. | | Deposit date: | 2022-11-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | BAY-7081: A Potent, Selective, and Orally Bioavailable Cyanopyridone-Based PDE9A Inhibitor.
J.Med.Chem., 65, 2022
|
|
5MQ1
 
 | | Crystal structure of the BRD7 bromodomain in complex with BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7, ... | | Authors: | Diaz-Saez, L, Martin, L.J, Panagakou, I, Picaud, S, Krojer, T, von Delft, F, Knapp, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the BRD7 bromodomain in complex with BI-9564
To Be Published
|
|
6XOY
 
 | |
6BEB
 
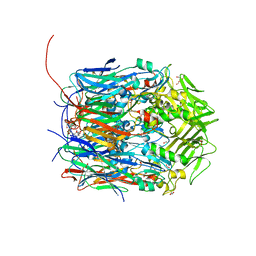 | | Crystal structure of VACV D13 in complex with Rifamycin SV | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Scaffold protein D13, ... | | Authors: | Garriga, D, Accurso, C, Coulibaly, F. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GJL
 
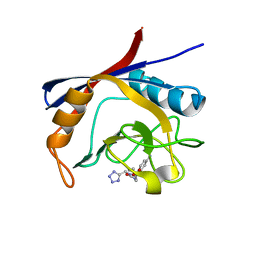 | | Cyclophilin A complexed with tri-vector ligand 10. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-(1~{H}-1,2,3,4-tetrazol-5-ylmethyl)carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
8DUJ
 
 | |
9EW1
 
 | | Ternary structure of 14-3-3s, CRAF phosphopeptide (pS259) and compound 79 (1124379). | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-1-[8-(4-iodophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]ethanone, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
4REZ
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease | | Descriptor: | ORF1ab protein, S-1,2-PROPANEDIOL, ZINC ION | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
3IG7
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFP with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-{1-[cis-3-(acetylamino)cyclobutyl]-1H-imidazol-4-yl}-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7Q1D
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
6F6U
 
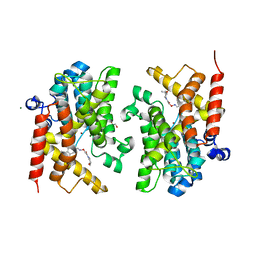 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-7b | | Descriptor: | 2-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-1-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
9ATV
 
 | | X-ray co-crystal structure of NCGC00685960 / NCATS-SM9335 in NNMT | | Descriptor: | (8M)-8-{3-[(3S)-1-(2-cyclohexylethyl)piperidin-3-yl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}quinolin-2(1H)-one, Nicotinamide N-methyltransferase | | Authors: | Olland, A, White, A, Suto, R. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | NNMT inhibition in cancer-associated fibroblasts restores antitumour immunity.
Nature, 645, 2025
|
|
8G74
 
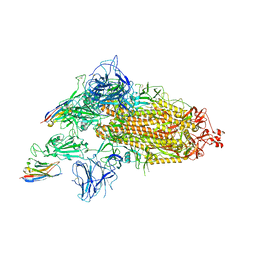 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
5O0U
 
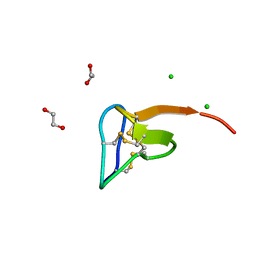 | | Crystal structure of tarantula venom peptide Protoxin-II | | Descriptor: | 1,2-ETHANEDIOL, Beta/omega-theraphotoxin-Tp2a, CHLORIDE ION | | Authors: | Tabor, A, McCarthy, S, Reyes, F.E. | | Deposit date: | 2017-05-17 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | The Role of Disulfide Bond Replacements in Analogues of the Tarantula Toxin ProTx-II and Their Effects on Inhibition of the Voltage-Gated Sodium Ion Channel Nav1.7.
J.Am.Chem.Soc., 139, 2017
|
|
4R5Z
 
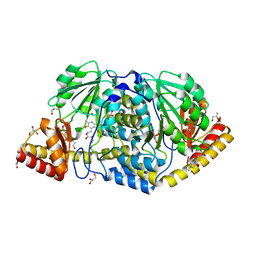 | | Crystal structure of Rv3772 encoded aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
6F90
 
 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
