6G2D
 
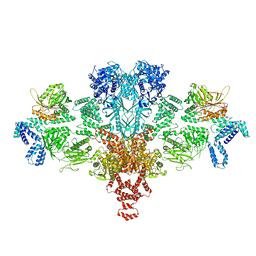 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
7X6F
 
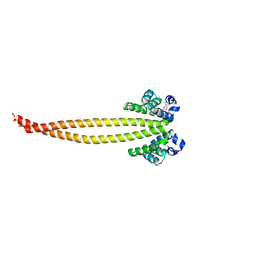 | | Outer membrane lipoprotein QseG of Escherichia coli O157:H7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CITRATE ANION, ... | | Authors: | Matsumoto, K, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-03-07 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6P40
 
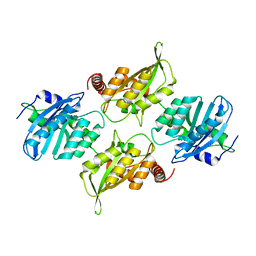 | | Crystal Structure of Full Length APOBEC3G FKL | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
9FF5
 
 | | The structure of G.kaustophilus T-1 ScoC-23bp dsDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*AP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*AP*AP*AP*AP*TP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*AP*AP*TP*AP*TP*TP*AP*TP*TP*TP*TP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*T)-3'), ... | | Authors: | Hadad, N, Shulami, S, Pomyalov, S, Shoham, Y, Shoham, G. | | Deposit date: | 2024-05-22 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of G.kaustophilus T-1 ScoC-23bp dsDNA complex
To be Published
|
|
7ZIB
 
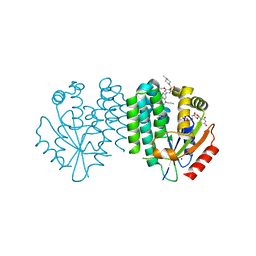 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0635 inhibitor | | Descriptor: | 2-[2-[[5-[3-methoxy-4-(4-methylpiperazin-1-yl)sulfonyl-phenyl]-2-methyl-phenyl]-propyl-amino]-1,3-thiazol-4-yl]pyrimidine-4,6-diamine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Ben-Yaala, K, Saez-Ayala, M, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
6RTX
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
5INK
 
 | |
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
4XHX
 
 | |
5IOY
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)pyrrolidine-1-carboxamide at 1.77A resolution | | Descriptor: | N-(cyclopentylmethyl)pyrrolidine-1-carboxamide, SULFATE ION, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
6RWC
 
 | |
4Q0H
 
 | | Deinococcus radiodurans BphP PAS-GAF | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
7X9D
 
 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
5I2L
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
6DHM
 
 | | Bovine glutamate dehydrogenase complexed with zinc | | Descriptor: | GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
6P0R
 
 | | Methyltransferase domain of human suppressor of variegation 3-9 homolog 2 (SUV39H2) in complex with OTS186935 inhibitor | | Descriptor: | (3S)-1-[2-(5-chloro-2,4-dimethoxyphenyl)imidazo[1,2-a]pyridin-7-yl]-N-[(pyridin-4-yl)methyl]pyrrolidin-3-amine, Histone-lysine N-methyltransferase SUV39H2, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methyltransferase domain of human suppressor of variegation 3-9 homolog 2 (SUV39H2) in complex with OTS186935 inhibitor
to be published
|
|
4Q2J
 
 | | A novel structure-based mechanism for DNA-binding of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Wang, Z, Long, J, Yang, X, Shen, Y. | | Deposit date: | 2014-04-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Crystal structure of the ubiquitin-like domain-CUT repeat-like tandem of special AT-rich sequence binding protein 1 (SATB1) reveals a coordinating DNA-binding mechanism.
J.Biol.Chem., 289, 2014
|
|
6P25
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
8THH
 
 | | Cryo-EM structure of Nav1.7 with LTG | | Descriptor: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2ROU
 
 | | Stereospecific Conformations of N2-dG 1R-trans-anti-Benzo[c]phenanthrene DNA Adducts: 3'-Intercalation of the 1R Adduct and 5'-Minor Groove Orientation of the 1S Adduct in an Iterated (CG)3 Repeat | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, DNA (5'-D(*DAP*DTP*DCP*DGP*DCP*DGP*DCP*DGP*DGP*DCP*DAP*DTP*DG)-3'), DNA (5'-D(*DCP*DAP*DTP*DGP*DCP*DCP*DGP*DCP*DGP*DCP*DGP*DAP*DT)-3') | | Authors: | Wang, Y, Kroth, H, Yagi, H, Sayer, J.M, Kumar, S, Jerina, D.M, Stone, M.P. | | Deposit date: | 2008-04-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 3'-Intercalation of a N2-dG 1R-trans-anti-benzo[c]phenanthrene DNA adduct in an iterated (CG)3 repeat
Chem.Res.Toxicol., 21, 2008
|
|
9JI0
 
 | | 3-Hydroxybutyryl-CoA dehydrogenase | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain protein | | Authors: | Yang, J.W, Jeon, H.J, Park, S.H, Jang, S.H, Park, J.A, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Insights and Catalytic Mechanism of 3-Hydroxybutyryl-CoA Dehydrogenase from Faecalibacterium Prausnitzii A2-165.
Int J Mol Sci, 25, 2024
|
|
