9EQ5
 
 | |
6RES
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9HHW
 
 | | Crystal structure of TTBK1 with a covalent compound GCL95 | | Descriptor: | Tau-tubulin kinase 1, ~{N}-[2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)phenyl]propanamide | | Authors: | Wang, G, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-22 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
6R4B
 
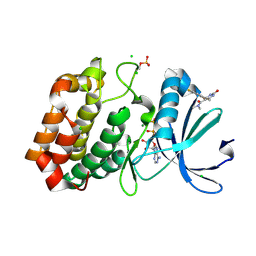 | | Aurora-A in complex with shape-diverse fragment 56 | | Descriptor: | (6~{S})-6-[2,4-bis(fluoranyl)phenyl]-~{N},~{N},4-trimethyl-2-oxidanylidene-5,6-dihydro-1~{H}-pyrimidine-5-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
7T0C
 
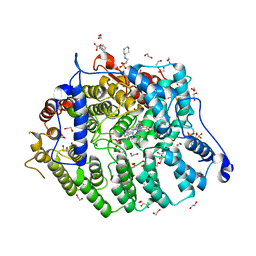 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2e | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-5-butyl-4-({1-[(4-chlorophenyl)methyl]-1H-imidazol-5-yl}methyl)-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
6YK6
 
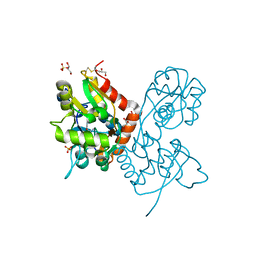 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound (S)-1-(2'-Amino-2'-carboxyethyl)furo[3,4-d]pyrimidin-2,4-dione at resolution 1.47A | | Descriptor: | (S)-1-(2'-Amino-2'-carboxyethyl)furo[3,4-d]pyrimidin-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
9BR4
 
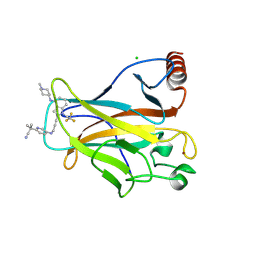 | | Crystal structure of p53 Y220C mutant in complex with PC-9859 | | Descriptor: | 2-methyl-2-{5-[(3-{4-[(1-methylpiperidin-4-yl)amino]-1-(2,2,2-trifluoroethyl)-1H-indol-2-yl}prop-2-yn-1-yl)amino]pyridin-2-yl}propanenitrile, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Abendroth, J, Lorimer, D.D, Vu, B, Takana, N. | | Deposit date: | 2024-05-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Rezatapopt (PC14586), a First-in-Class, Small-Molecule Reactivator of p53 Y220C Mutant in Development.
Acs Med.Chem.Lett., 16, 2025
|
|
6RBV
 
 | |
5L1A
 
 | | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila
To Be Published
|
|
6YEO
 
 | | Crystal structure of AmpC from E. coli with cyclic boronate 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Lang, P.A, Schofield, C.J, Brem, J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
6YGZ
 
 | | tRNA Guanine Transglycosylase (TGT) labelled with 5-fluoro-tryptophan in co-crystallized complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Co-crystallization, 19F NMR and nanoESI-MS reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6GNN
 
 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
7XJ9
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab | | Descriptor: | 55A8 heavy chain, 55A8 light chain, 58G6 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab
To Be Published
|
|
7BF1
 
 | | Ca2+-Calmodulin in complex with peptide from brain-type creatine kinase in extended 1:2 binding mode | | Descriptor: | ACETYL GROUP, CALCIUM ION, Calmodulin-1, ... | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
7T6D
 
 | | CryoEM structure of the YejM/LapB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Inner membrane protein YejM, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli.
Nat Commun, 13, 2022
|
|
4UAF
 
 | |
6RE2
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9MWZ
 
 | | Cryo-EM Structure of Human Enterovirus D68 USA/IL/14-18952 | | Descriptor: | viral protein 1, viral protein 2, viral protein 3, ... | | Authors: | Xu, L, Pintilie, G, Varanese, L, Carette, J.E, Chiu, W. | | Deposit date: | 2025-01-17 | | Release date: | 2025-03-26 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | MFSD6 is an entry receptor for enterovirus D68.
Nature, 641, 2025
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8Y6J
 
 | | The structure of Oryza sativa HKT1;1 | | Descriptor: | Cation transporter HKT1;1,Soluble cytochrome b562 | | Authors: | Yang, G.H, Gao, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural insights into the Oryza sativa cation transporters HKTs in salt tolerance.
J Integr Plant Biol, 66, 2024
|
|
9BXL
 
 | | OvoM from Sulfuricurvum sp. isolate STB_99, an ovothiol-biosynthetic N-methyltransferase in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-thiohistidine N-methyltransferase OvoM, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-05-22 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and functional analysis of SAM-dependent N-methyltransferases involved in ovoselenol and ovothiol biosynthesis.
Structure, 33, 2025
|
|
7XEZ
 
 | |
8Y6M
 
 | | The structure of Oryza sativa HKT2;1 | | Descriptor: | Cation transporter HKT2;1,Soluble cytochrome b562 | | Authors: | Yang, G.H, Gao, R, Jia, Y.T, Xu, X, Fu, P, Zhou, J.Q. | | Deposit date: | 2024-02-02 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into the Oryza sativa cation transporters HKTs in salt tolerance.
J Integr Plant Biol, 66, 2024
|
|
